8RF0
 
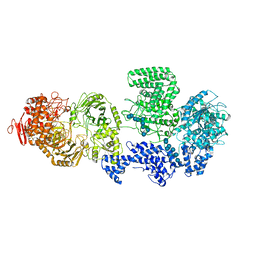 | | WT-CGS sample in nanodisc | | Descriptor: | Cyclic beta-(1,2)-glucan synthase NdvB, URIDINE-5'-DIPHOSPHATE-GLUCOSE, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
8RF9
 
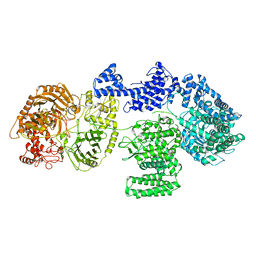 | | CgsiGP1 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
8RFE
 
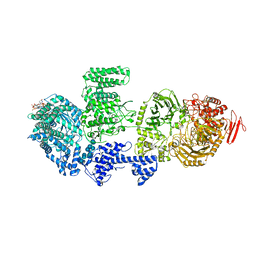 | | CgsiGP2 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, URIDINE-5'-DIPHOSPHATE-GLUCOSE, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
8RFG
 
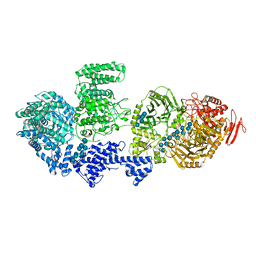 | | CgsiGP3 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
7ZBR
 
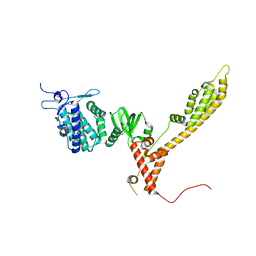 | | Crystal Structure of full-length Bartonella Effector protein 1 | | Descriptor: | Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Huber, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length structure of a FIC effector protein from Bartonella reveals a novel structural domain
To be published
|
|
7ZO8
 
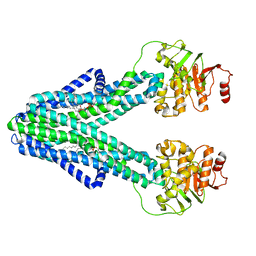 | | cryo-EM structure of CGT ABC transporter in nanodisc apo state | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, DIUNDECYL PHOSPHATIDYL CHOLINE | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZOA
 
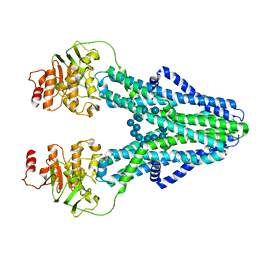 | | cryo-EM structure of CGT ABC transporter in presence of CBG substrate | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, Cyclooctadecakis-(1-2)-(beta-D-glucopyranose) | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZO9
 
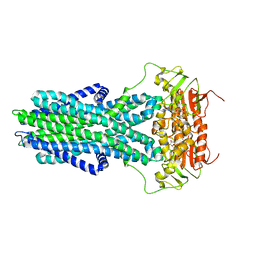 | | cryo-EM structure of CGT ABC transporter in vanadate trapped state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZNU
 
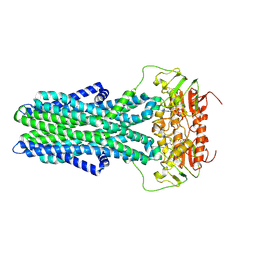 | | cryo-EM structure of CGT ABC transporter in detergent micelle | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2JK8
 
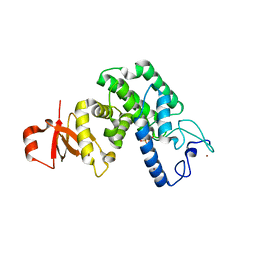 | |
2VY3
 
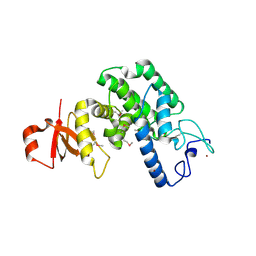 | | Type IV secretion system effector protein BepA | | Descriptor: | CELL FILAMENTATION PROTEIN, CHLORIDE ION, NICKEL (II) ION | | Authors: | Palanivelu, D.V, Schirmer, T. | | Deposit date: | 2008-07-17 | | Release date: | 2009-11-17 | | Last modified: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fic Domain Catalyzed Adenylylation: Insight Provided by the Structural Analysis of the Type Iv Secretion System Effector Bepa.
Protein Sci., 20, 2011
|
|
2VZA
 
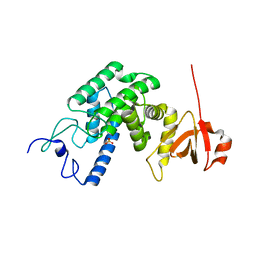 | | Type IV secretion system effector protein BepA | | Descriptor: | CELL FILAMENTATION PROTEIN, SULFATE ION | | Authors: | Meury, M, Schirmer, T. | | Deposit date: | 2008-07-31 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Fic Domain Catalyzed Adenylylation: Insight Provided by the Structural Analysis of the Type Iv Secretion System Effector Bepa.
Protein Sci., 20, 2011
|
|
4WGJ
 
 | |
4XI8
 
 | |
4YK2
 
 | |
4YK1
 
 | |
4YK3
 
 | |
4M16
 
 | |
4LU4
 
 | |
4N67
 
 | |
5EU0
 
 | |
5CMT
 
 | |
5CGL
 
 | |
5CKL
 
 | |
5JFF
 
 | | E. coli EcFicT mutant G55R in complex with EcFicA | | Descriptor: | CHLORIDE ION, Probable adenosine monophosphate-protein transferase fic, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | Authors: | Stanger, F.V, Schirmer, T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Escherichia coli Fic Toxin-Like Protein in Complex with Its Cognate Antitoxin.
Plos One, 11, 2016
|
|
