7WXL
 
 | |
7WXK
 
 | |
7WXQ
 
 | |
7WXN
 
 | |
7WXJ
 
 | |
7WXR
 
 | |
7WXM
 
 | |
7WXO
 
 | |
7WXP
 
 | |
7XTF
 
 | |
7XTE
 
 | |
6ICE
 
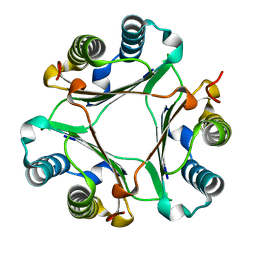 | | Crystal structure of Hamster MIF | | Descriptor: | Macrophage migration inhibitory factor | | Authors: | Sundaram, R, Vasudevan, D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Macrophage migration inhibitory factor of Syrian golden hamster shares structural and functional similarity with human counterpart and promotes pancreatic cancer.
Sci Rep, 9, 2019
|
|
