5COK
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COO
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
4PGK
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, FE (III) ION | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PGI
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Analogs Bound | | Descriptor: | 1,2-ETHANEDIOL, 11-[2-(2-ethoxyethoxy)ethoxy]undecanal, Aldehyde decarbonylase, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-02 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PG1
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | (1S,2S)-2-nonylcyclopropanecarboxylic acid, Aldehyde decarbonylase, DIMETHYL SULFOXIDE, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4PG0
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | (1S,2S)-2-nonylcyclopropanecarboxylic acid, Aldehyde decarbonylase, DIMETHYL SULFOXIDE, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-05-01 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4RVJ
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with amprenavir | | Descriptor: | HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2014-11-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1
To be Published
|
|
4RVI
 
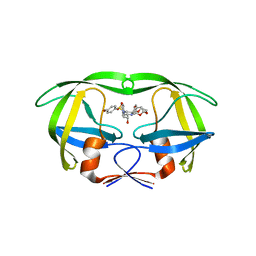 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL0519 | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2014-11-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1
To be Published
|
|
4RVX
 
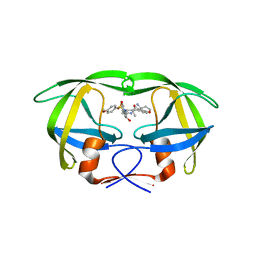 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL079 | | Descriptor: | (4S)-4-amino-N-[(2S,3S)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-3,4-dihydro-2H-chromene-6-carboxamide, HIV-1 Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2014-11-28 | | Release date: | 2016-05-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1.
To be Published
|
|
4TW3
 
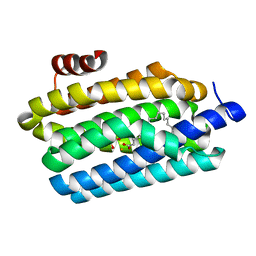 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
5TYR
 
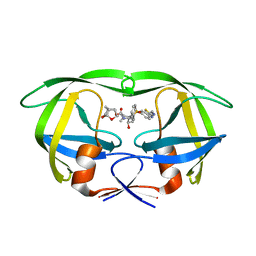 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
5TYS
 
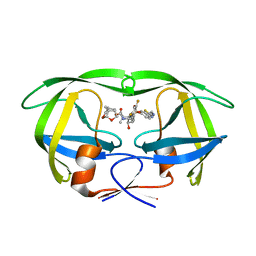 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | A novel central nervous system-penetrating protease inhibitor overcomes human immunodeficiency virus 1 resistance with unprecedented aM to pM potency.
Elife, 6, 2017
|
|
7JKV
 
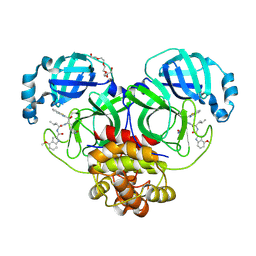 | | Crystal Structure of SARS-CoV-2 main protease in complex with an inhibitor GRL-2420 | | Descriptor: | 3C-like proteinase, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, PENTAETHYLENE GLYCOL | | Authors: | Bulut, H, Hattori, S.I, Das, D, Murayama, K, Mitsuya, H. | | Deposit date: | 2020-07-29 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication.
Nat Commun, 12, 2021
|
|
4HLA
 
 | | Crystal structure of wild type HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-10-16 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
4I8W
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL007 | | Descriptor: | 4-{[(2R,3S)-3-({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yloxy]carbonyl}amino)-2-hydroxy-4-phenylbutyl](2-methylpropyl)sulfamoyl}benzoic acid, Protease | | Authors: | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
4I8Z
 
 | | Crystal structure of wild type HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Palmer, I, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel non-peptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety.
Antimicrob.Agents Chemother., 57, 2013
|
|
4NJS
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with non-peptidic inhibitor, GRL008 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-carbamoylphenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJT
 
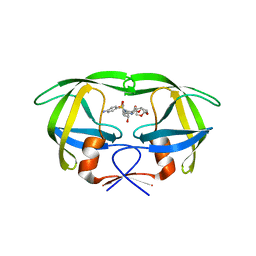 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJU
 
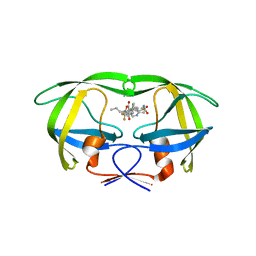 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with tipranavir | | Descriptor: | N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, Protease | | Authors: | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
4NJV
 
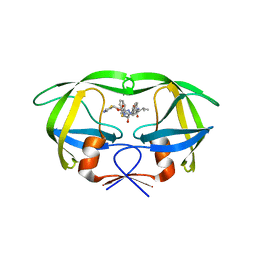 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with ritonavir | | Descriptor: | Protease, RITONAVIR | | Authors: | Yedidi, R.S, Garimella, H, Chang, S.B, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Hydrogen-Bonding Network of P2 bis-Tetrahydrofuran-Containing HIV-1 Protease Inhibitors (PIs) with a Protease Active-Site Amino Acid Backbone Aids in Their Activity against PI-Resistant HIV.
Antimicrob.Agents Chemother., 58, 2014
|
|
3BY7
 
 | |
3DEE
 
 | |
2Q8U
 
 | |
3G23
 
 | |
