2UV1
 
 | |
2UUZ
 
 | |
1W5S
 
 | | Structure of the Aeropyrum Pernix ORC2 protein (ADP form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ORIGIN RECOGNITION COMPLEX SUBUNIT 2 ORC2, SULFATE ION | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
1W5T
 
 | | Structure of the Aeropyrum Pernix ORC2 protein (ADPNP-ADP complexes) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ORC2, ... | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
2CHV
 
 | | Replication Factor C ADPNP complex | | Descriptor: | REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2CHQ
 
 | | Replication Factor C ADPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2CHG
 
 | | Replication Factor C domains 1 and 2 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
3GPL
 
 | | Crystal structure of the ternary complex of RecD2 with DNA and ADPNP | | Descriptor: | 5'-D(*T*TP*TP*TP*TP*TP*TP*T)-3', Exodeoxyribonuclease V, subunit RecD, ... | | Authors: | Saikrishnan, K, Cook, N, Wigley, D.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of 5'-3' translocation in SF1B helicases.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GP8
 
 | | Crystal structure of the binary complex of RecD2 with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*T*TP*TP*TP*TP*TP*TP*TP*T)-3', Exodeoxyribonuclease V, subunit RecD, ... | | Authors: | Saikrishnan, K, Cook, N, Wigley, D.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic basis of 5'-3' translocation in SF1B helicases.
Cell(Cambridge,Mass.), 137, 2009
|
|
3E1S
 
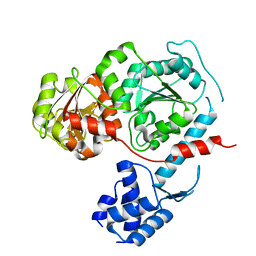 | | Structure of an N-terminal truncation of Deinococcus radiodurans RecD2 | | Descriptor: | Exodeoxyribonuclease V, subunit RecD | | Authors: | Saikrishnan, K, Griffiths, S.P, Cook, N, Court, R, Wigley, D.B. | | Deposit date: | 2008-08-04 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding to RecD: role of the 1B domain in SF1B helicase activity.
Embo J., 27, 2008
|
|
5CZ1
 
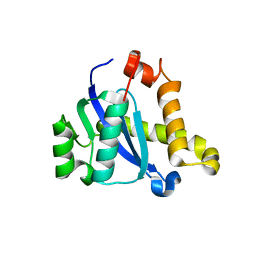 | |
5CZ2
 
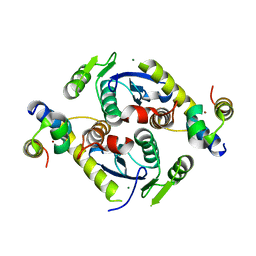 | | Crystal structure of a two-domain fragment of MMTV integrase | | Descriptor: | MAGNESIUM ION, Pol polyprotein, ZINC ION | | Authors: | Cook, N, Ballandras-Colas, A, Engelman, A, Cherepanov, P. | | Deposit date: | 2015-07-31 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
6GX9
 
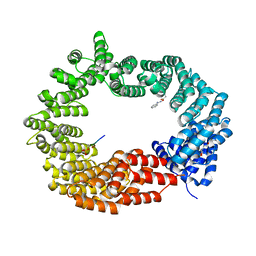 | | Crystal structure of the TNPO3 - CPSF6 RSLD complex | | Descriptor: | BENZAMIDINE, BICINE, Cleavage and polyadenylation specificity factor subunit 6, ... | | Authors: | Cherepanov, P, Cook, N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6.
Nucleic Acids Res., 47, 2019
|
|
6RWO
 
 | | SIVrcm intasome (Q148H/G140S) in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWL
 
 | | SIVrcm intasome | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP*CP*A)-3'), Pol protein, ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWM
 
 | | SIVrcm intasome in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
3IW4
 
 | | Crystal structure of PKC alpha in complex with NVP-AEB071 | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]-1H-pyrrole-2,5-dione, Protein kinase C alpha type | | Authors: | Stark, W, Rummel, G, Strauss, A, Cowan-Jacob, S.W. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(1H-indol-3-yl)-4-[2-(4-methylpiperazin-1-yl)quinazolin-4-yl]pyrrole-2,5-dione (AEB071), a potent and selective inhibitor of protein kinase C isotypes
J.Med.Chem., 52, 2009
|
|
3K70
 
 | |
