2M8N
 
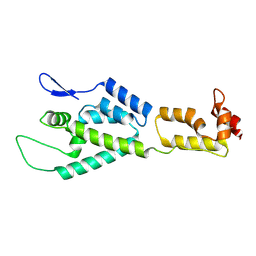 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M8P
 
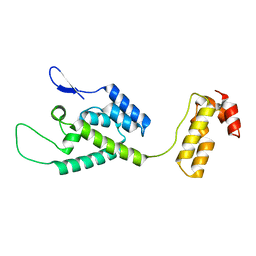 | | The structure of the W184AM185A mutant of the HIV-1 capsid protein | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M8L
 
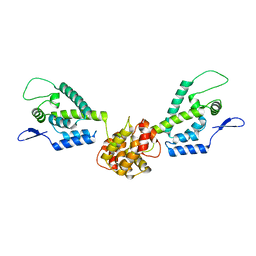 | | HIV capsid dimer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-23 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2N5T
 
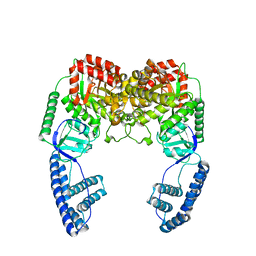 | | Ensemble solution structure of the phosphoenolpyruvate-Enzyme I complex from the bacterial phosphotransferase system | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Venditti, V, Schwieters, C.D, Grishaev, A, Clore, G. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-02 | | Last modified: | 2019-04-17 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Dynamic equilibrium between closed and partially closed states of the bacterial Enzyme I unveiled by solution NMR and X-ray scattering.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2L5H
 
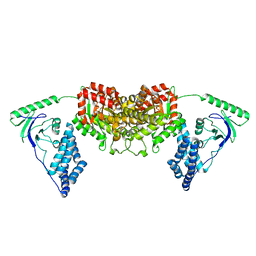 | | Solution Structure of the H189Q mutant of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Takayama, Y.D, Schwieters, C.D, Grishaev, A, Guirlando, R, Clore, G. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2012-04-25 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Combined Use of Residual Dipolar Couplings and Solution X-ray Scattering To Rapidly Probe Rigid-Body Conformational Transitions in a Non-phosphorylatable Active-Site Mutant of the 128 kDa Enzyme I Dimer.
J.Am.Chem.Soc., 133, 2011
|
|
1ZXJ
 
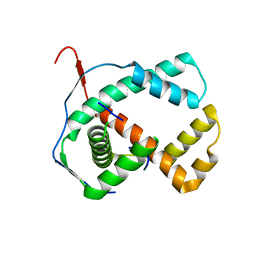 | | Crystal structure of the hypthetical Mycoplasma protein, MPN555 | | Descriptor: | Hypothetical protein MG377 homolog | | Authors: | Schulze-Gahmen, U, Aono, S, Shengfeng, C, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-06-08 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the hypothetical Mycoplasma protein MPN555 suggests a chaperone function.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2CI2
 
 | |
1T96
 
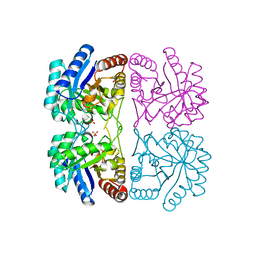 | | r106g kdo8ps with pep | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Gatti, D.L. | | Deposit date: | 2004-05-14 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Effects of the Arg106==>Gly mutation on the catalytic and conformational cycle of Aquifex aeolicus KDO8P synthase.
To be Published
|
|
1T99
 
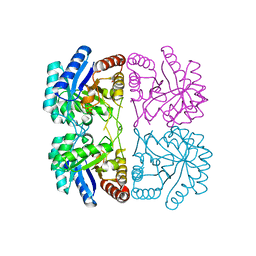 | | r106g kdo8ps without substrates | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION | | Authors: | Gatti, D.L. | | Deposit date: | 2004-05-16 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Effects of the Arg106==>Gly mutation on the catalytic and conformational cycle of Aquifex aeolicus KDO8P synthase.
To be Published
|
|
1T8X
 
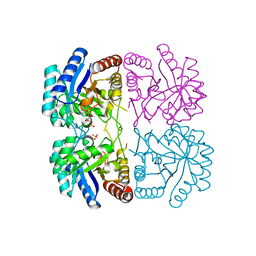 | | r106g kdo8ps with pep and a5p | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, CADMIUM ION, ... | | Authors: | Gatti, D.L. | | Deposit date: | 2004-05-13 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of the Arg106==>Gly mutation on the catalytic and conformational cycle of Aquifex aeolicus KDO8P synthase.
To be Published
|
|
1SXL
 
 | |
1ICW
 
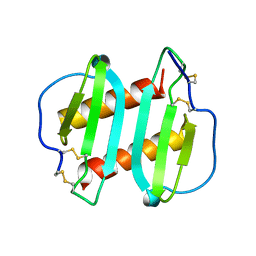 | | INTERLEUKIN-8, MUTANT WITH GLU 38 REPLACED BY CYS AND CYS 50 REPLACED BY ALA | | Descriptor: | INTERLEUKIN-8 | | Authors: | Eigenbrot, C, Lowman, H.B, Chee, L, Artis, D.R. | | Deposit date: | 1996-09-18 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural change and receptor binding in a chemokine mutant with a rearranged disulfide: X-ray structure of E38C/C50AIL-8 at 2 A resolution.
Proteins, 27, 1997
|
|
1HIC
 
 | |
1ILQ
 
 | |
1TVX
 
 | |
1UL9
 
 | | CGL2 ligandfree | | Descriptor: | galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULC
 
 | | CGL2 in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULF
 
 | | CGL2 in complex with Blood Group A tetrasaccharide | | Descriptor: | alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULE
 
 | | CGL2 in complex with linear B2 trisaccharide | | Descriptor: | alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULD
 
 | | CGL2 in complex with blood group H type II | | Descriptor: | alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ULG
 
 | | CGL2 in complex with Thomsen-Friedenreich antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, galectin-2 | | Authors: | Walser, P.J, Haebel, P.W, Kuenzler, M, Kues, U, Aebi, M, Ban, N. | | Deposit date: | 2003-09-12 | | Release date: | 2004-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Functional Analysis of the Fungal Galectin CGL2
STRUCTURE, 12, 2004
|
|
1ILP
 
 | |
1JE4
 
 | |
1WS1
 
 | |
2JZO
 
 | |
