1EZB
 
 | |
2XDF
 
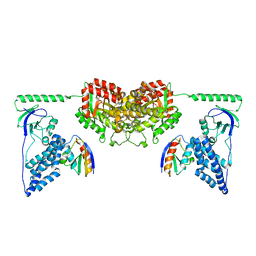 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2019-08-21 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2YHH
 
 | | Microvirin:mannobiose complex | | Descriptor: | MANNAN-BINDING LECTIN, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hussan, S, Bewley, C.A, Clore, G.M, Gustchina, E, Ghirlando, R. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Monovalent Lectin Microvirin in Complex with Man(Alpha)(1-2)Man Provides a Basis for Anti-HIV Activity with Low Toxicity.
J.Biol.Chem., 286, 2011
|
|
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
1VRV
 
 | |
1WJE
 
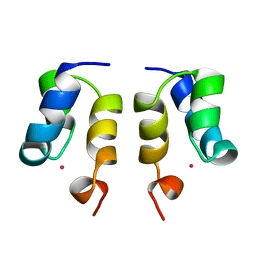 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
1ZHS
 
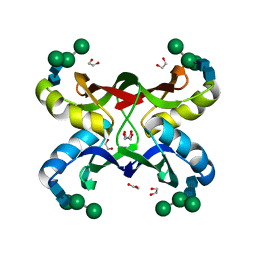 | | Crystal structure of MVL bound to Man3GlcNAc2 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
1ZHQ
 
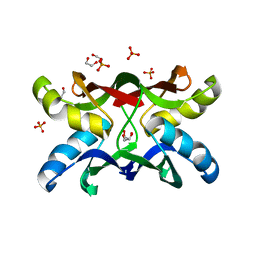 | | Crystal structure of apo MVL | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, mannan-binding lectin | | Authors: | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
1IOB
 
 | |
1WCR
 
 | |
7JSQ
 
 | | Refined structure of the C-terminal domain of DNAJB6b | | Descriptor: | DnaJ homolog subfamily B member 6 | | Authors: | Karamanos, T.K, Clore, G.M. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | An S/T motif controls reversible oligomerization of the Hsp40 chaperone DNAJB6b through subtle reorganization of a beta sheet backbone.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2GB1
 
 | |
2LV6
 
 | | The complex between Ca-Calmodulin and skeletal muscle myosin light chain kinase from combination of NMR and aqueous and contrast-matched SAXS data | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase 2, ... | | Authors: | Grishaev, A.V, Anthis, N.J, Clore, G.M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-20 | | Last modified: | 2013-03-27 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Contrast-matched small-angle X-ray scattering from a heavy-atom-labeled protein in structure determination: application to a lead-substituted calmodulin-peptide complex.
J.Am.Chem.Soc., 134, 2012
|
|
2EZP
 
 | |
2EZS
 
 | |
2EZR
 
 | |
2EZO
 
 | |
2EZQ
 
 | |
2EZN
 
 | |
2EZM
 
 | |
2NEF
 
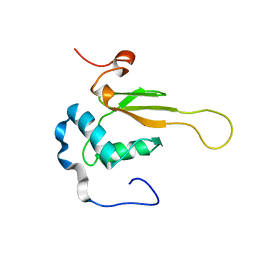 | | HIV-1 NEF (REGULATORY FACTOR), NMR, 40 STRUCTURES | | Descriptor: | NEGATIVE FACTOR (F-PROTEIN) | | Authors: | Grzesiek, S, Bax, A, Clore, G.M, Gronenborn, A.M, Hu, J.S, Kaufman, J, Palmer, I, Stahl, S.J, Tjandra, N, Wingfield, P.T. | | Deposit date: | 1997-02-12 | | Release date: | 1997-07-07 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure and backbone dynamics of HIV-1 Nef.
Protein Sci., 6, 1997
|
|
3MAC
 
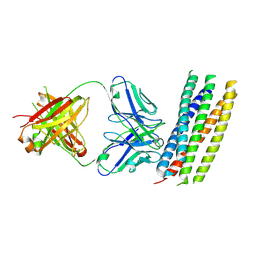 | | crystal structure of GP41-derived protein complexed with fab 8062 | | Descriptor: | Fab8062, Transmembrane glycoprotein | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
3MA9
 
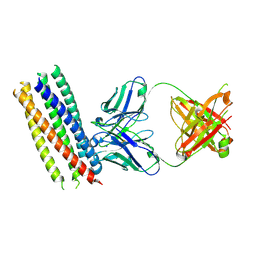 | | Crystal structure of gp41 derived protein complexed with fab 8066 | | Descriptor: | Fab8066 FAB ANTIBODY FRAGMENT, Heavy Chain, Light Chain, ... | | Authors: | Li, M, Gustchina, E, Louis, J, Gustchina, A, Wlodawer, A, Clore, M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of HIV-1 Neutralization by Affinity Matured Fabs Directed against the Internal Trimeric Coiled-Coil of gp41.
Plos Pathog., 6, 2010
|
|
4KHX
 
 | |
3CI2
 
 | |
