4PFB
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Fusobacterium nucleatun (FN1258, TARGET EFI-510120) with bound SN-glycerol-3-phosphate | | Descriptor: | C4-dicarboxylate-binding protein, IMIDAZOLE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PF6
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS (RD1_0742, TARGET EFI-510239) WITH BOUND 3-DEOXY-D-MANNO-OCT-2-ULOSONIC ACID (KDO) | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, C4-dicarboxylate-binding protein, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PGN
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND INDOLE PYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PGP
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND 3-INDOLE ACETIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PET
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM COLWELLIA PSYCHRERYTHRAEA (CPS_0129, TARGET EFI-510097) WITH BOUND CALCIUM AND PYRUVATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3TS7
 
 | | CRYSTAL STRUCTURE OF FARNESYL DIPHOSPHATE SYNTHASE (TARGET EFI-501951) FROM Methylococcus capsulatus | | Descriptor: | Geranyltranstransferase, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UCA
 
 | | Crystal structure of isoprenoid synthase (target EFI-501974) from clostridium perfringens | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-26 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KKN
 
 | | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 | | Descriptor: | Cytotoxic T-lymphocyte associated protein 4, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Bonanno, J, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Scott Glenn, A, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704
to be published
|
|
4KNP
 
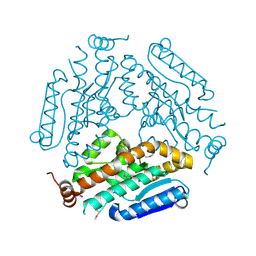 | | Crystal Structure Of a Putative enoyl-coA hydratase (PSI-NYSGRC-019597) from Mycobacterium avium paratuberculosis K-10 | | Descriptor: | enoyl-CoA hydratase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-10 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of a Putative enoyl-coA hydratase from Mycobacterium avium paratuberculosis K-10
to be published
|
|
4KTO
 
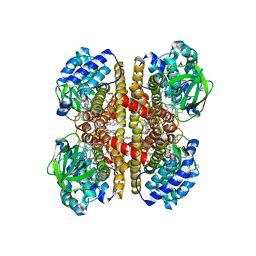 | | Crystal Structure Of a Putative Isovaleryl-CoA dehydrogenase (PSI-NYSGRC-012251) from Sinorhizobium meliloti 1021 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, isovaleryl-CoA dehydrogenase | | Authors: | Kumar, P.R, Ahmed, M, Attonito, J, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Glenn, A.S, Hammonds, J, Hillerich, B, Himmel, D, Love, J.D, Seidel, R, Stead, M, Toro, R, Wasserman, S.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Crystal structure of a Putative Isovaleryl-CoA dehydrogenase from Sinorhizobium meliloti 1021
to be published
|
|
4N8P
 
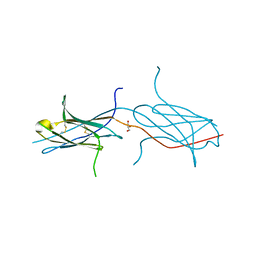 | | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Uncharacterized protein | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of a strand swapped CTLA-4 from Duckbill Platypus [PSI-NYSGRC-012711]
to be published
|
|
4N68
 
 | | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804] | | Descriptor: | Contactin-5, SULFATE ION | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an internal FN3 domain from human Contactin-5 [PSI-NYSGRC-005804]
to be published
|
|
4N5U
 
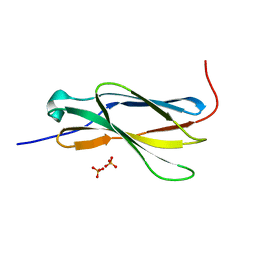 | | Crystal structure of the 4th FN3 domain of human Protein Tyrosine phosphatase, receptor type F [PSI-NYSGRC-006240] | | Descriptor: | Receptor-type tyrosine-protein phosphatase F, SULFATE ION | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-10-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.456 Å) | | Cite: | Crystal structure of the 4th FN3 domain of human PTP, receptor F [PSI-NYSGRC-006240]
to be published
|
|
4NOF
 
 | | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Polymeric immunoglobulin receptor | | Authors: | Sampathkumar, P, Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the second Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4NOB
 
 | | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor [PSI-NYSGRC-006220] | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Kumar, P.R, Banu, R, Bhosle, R, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Glenn, A.S, Hillerich, B, Khafizov, K, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of the 1st Ig domain from mouse Polymeric Immunoglobulin receptor
to be published
|
|
4RS3
 
 | | Crystal structure of carbohydrate transporter A0QYB3 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with xylitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4RSM
 
 | | Crystal structure of carbohydrate transporter msmeg_3599 from mycobacterium smegmatis str. mc2 155, target efi-510970, in complex with d-threitol | | Descriptor: | D-Threitol, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4DHD
 
 | | Crystal structure of isoprenoid synthase A3MSH1 (TARGET EFI-501992) from Pyrobaculum calidifontis | | Descriptor: | ACETATE ION, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FP4
 
 | | Crystal structure of isoprenoid synthase a3mx09 (target efi-501993) from pyrobaculum calidifontis | | Descriptor: | GERAN-8-YL GERAN, Polyprenyl synthetase, UNKNOWN ATOM OR ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F62
 
 | | Crystal structure of a putative farnesyl-diphosphate synthase from Marinomonas sp. MED121 (Target EFI-501980) | | Descriptor: | CHLORIDE ION, GLYCEROL, Geranyltranstransferase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
