6UEC
 
 | | Pseudomonas aeruginosa LpxD Complex Structure with Ligand | | Descriptor: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UED
 
 | | Apo Pseudomonas aeruginosa LpxD Structure | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
2L3N
 
 | | Solution structure of Rap1-Taz1 fusion protein | | Descriptor: | DNA-binding protein rap1,Telomere length regulator taz1 | | Authors: | Zhou, Z.R, Wang, F, Chen, Y, Lei, M, Hu, H. | | Deposit date: | 2010-09-19 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3R24
 
 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
7RBY
 
 | | Crystal structure of Nanobody nb112 and SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ilama-isolated nanobody NIH-CoV nb-112 specific to SARS-CoV-2 RBD, MAGNESIUM ION, ... | | Authors: | Chen, Y, Tolbert, W, Pazgier, M. | | Deposit date: | 2021-07-06 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Nebulized delivery of a broadly neutralizing SARS-CoV-2 RBD-specific nanobody prevents clinical, virological, and pathological disease in a Syrian hamster model of COVID-19.
Mabs, 14
|
|
7RQ6
 
 | | Cryo-EM structure of SARS-CoV-2 spike in complex with non-neutralizing NTD-directed CV3-13 Fab isolated from convalescent individual | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CV3-13 Fab heavy chain, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-08-05 | | Release date: | 2022-04-20 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection.
Cell Rep, 38, 2022
|
|
6J0Q
 
 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
3DB1
 
 | |
8CZZ
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with Temsavir, 8ANC195, and 10-1074 | | Descriptor: | 1-[4-(benzenecarbonyl)piperazin-1-yl]-2-[4-methoxy-7-(3-methyl-1H-1,2,4-triazol-1-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Pozharski, E, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-05-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
8DOK
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
5SY1
 
 | | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | | Descriptor: | CALCIUM ION, CHOLESTEROL, Calmodulin, ... | | Authors: | Clarke, O.B, Chen, Y, Mancia, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
8F93
 
 | | WDR5 covalently modified at Y228 by (R)-2-SF | | Descriptor: | 3-ethynyl-5-{[(3R)-4-{1-[(2-methoxyphenyl)methyl]-1H-benzimidazole-5-carbonyl}-3-methylpiperazin-1-yl]methyl}benzene-1-sulfonyl fluoride, CHLORIDE ION, GLYCEROL, ... | | Authors: | Taunton, J, Craven, G.B, Chen, Y. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes.
Nat.Chem., 15, 2023
|
|
8G6U
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CRF-1_AE T/F100 HIV-1 gp41, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
4C0E
 
 | | Structure of the NOT1 superfamily homology domain from Chaetomium thermophilum | | Descriptor: | NOT1 | | Authors: | Chen, Y, Boland, A, Raisch, T, Jonas, S, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2013-08-01 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Assembly of the not Module of the Human Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
4CRV
 
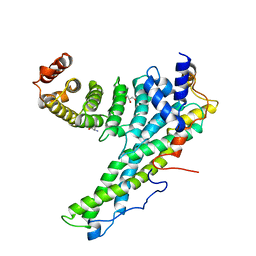 | | Complex of human CNOT9 and CNOT1 including two tryptophans | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG, GLYCEROL, ... | | Authors: | Boland, A, Chen, Y, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2014-03-01 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Ddx6-Cnot1 Complex and W-Binding Pockets in Cnot9 Reveal Direct Links between Mirna Target Recognition and Silencing
Mol.Cell, 54, 2014
|
|
2IKQ
 
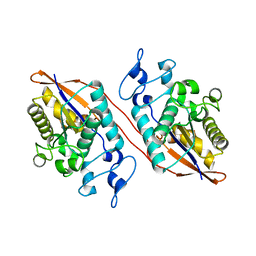 | |
2LS0
 
 | |
2N82
 
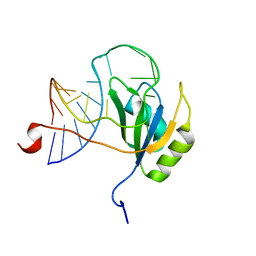 | |
2N7X
 
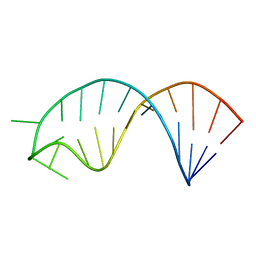 | | Solution structure of microRNA 20b pre-element | | Descriptor: | RNA (5'-R(*GP*GP*UP*AP*GP*UP*UP*UP*UP*GP*GP*CP*AP*UP*GP*AP*CP*UP*CP*UP*AP*CP*C)-3') | | Authors: | Yang, F, Chen, Y, Varani, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer.
Nucleic Acids Res., 44, 2016
|
|
3P2T
 
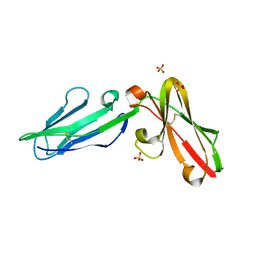 | | Crystal Structure of Leukocyte Ig-like Receptor LILRB4 (ILT3/LIR-5/CD85k) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, SULFATE ION | | Authors: | Chen, Y, Nam, G, Cheng, H, Zhang, J.H, Willcox, B.E, Gao, G.F. | | Deposit date: | 2010-10-04 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance
J.Biol.Chem., 286, 2011
|
|
3VAT
 
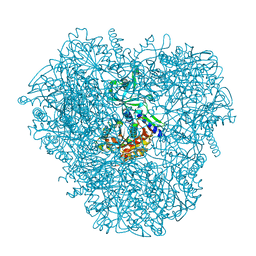 | | Crystal structure of DNPEP, ZnMg form | | Descriptor: | Aspartyl aminopeptidase, MAGNESIUM ION, ZINC ION | | Authors: | Kiser, P.D, Chen, Y, Palczewski, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into substrate specificity and metal activation of Mammalian tetrahedral aspartyl aminopeptidase.
J.Biol.Chem., 287, 2012
|
|
3VAR
 
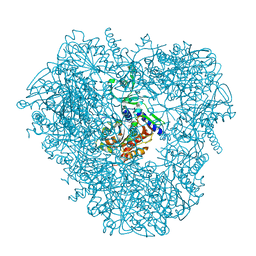 | | Crystal structure of DNPEP, ZnZn form | | Descriptor: | Aspartyl aminopeptidase, ZINC ION | | Authors: | Kiser, P.D, Chen, Y, Palczewski, K. | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into substrate specificity and metal activation of Mammalian tetrahedral aspartyl aminopeptidase.
J.Biol.Chem., 287, 2012
|
|
4IIK
 
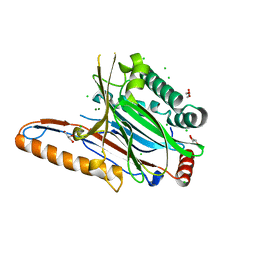 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
4IIP
 
 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
5YC2
 
 | |
