6KHD
 
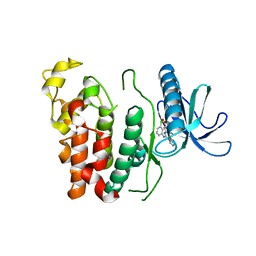 | | Crystal structure of CLK1 in complex with CX-4945 | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK1 | | Authors: | Lee, J.Y, Yun, J.S, Jin, H, Chang, J.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Selective Inhibition of Cdc2-Like Kinases by CX-4945.
Biomed Res Int, 2019, 2019
|
|
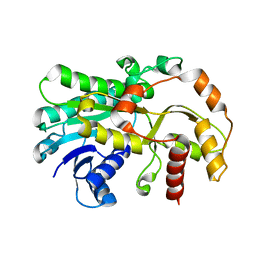
6KWS
 
 | |
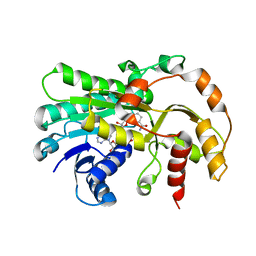
6KWT
 
 | |
4J7N
 
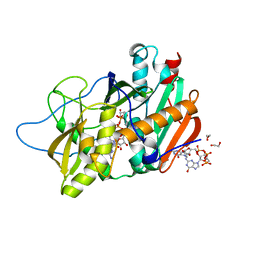 | | Crystal structure of mouse DXO in complex with M7GPPPG cap | | Descriptor: | 1,2-ETHANEDIOL, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 9-METHYLGUANINE, ... | | Authors: | Kilic, T, Chang, J.H, Tong, L. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A mammalian pre-mRNA 5' end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing.
Mol.Cell, 50, 2013
|
|
7E6H
 
 | | glucose-6-phosphate dehydrogenase from Kluyveromyces lactis | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Ha, V.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
7E6I
 
 | | Glucose-6-phosphate dehydrogenase in complex with its substrate glucose-6-phosphate | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Vu, H.H, Chang, J.H. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural basis for substrate recognition of glucose-6-phosphate dehydrogenase from Kluyveromyces lactis.
Biochem.Biophys.Res.Commun., 553, 2021
|
|
7ELF
 
 | |
4PZC
 
 | |
4PZE
 
 | |
4PZD
 
 | |
3PQ1
 
 | | Crystal structure of human mitochondrial poly(A) polymerase (PAPD1) | | Descriptor: | Poly(A) RNA polymerase | | Authors: | Bai, Y, Srivastava, S.K, Chang, J.H, Tong, L. | | Deposit date: | 2010-11-25 | | Release date: | 2011-03-30 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase.
Mol.Cell, 41, 2011
|
|
5ELM
 
 | | Crystal structure of L-aspartate/glutamate specific racemase in complex with L-glutamate | | Descriptor: | Asp/Glu_racemase family protein, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Ahn, J.W, Chang, J.H, Kim, K.J. | | Deposit date: | 2015-11-04 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for an atypical active site of an l-aspartate/glutamate-specific racemase from Escherichia coli
Febs Lett., 589, 2015
|
|
5ELL
 
 | |
1S28
 
 | | Crystal Structure of AvrPphF ORF1, the Chaperone for the Type III Effector AvrPphF ORF2 from P. syringae | | Descriptor: | ORF1, SULFATE ION | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.L, Sondek, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
1S21
 
 | | Crystal Structure of AvrPphF ORF2, A Type III Effector from P. syringae | | Descriptor: | ORF2 | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.K, Sondek, J. | | Deposit date: | 2004-01-07 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
5XR4
 
 | | Crystal structure of RabA1a in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a, ... | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
5XR7
 
 | | Crystal structure of RabA1a (Q72K) in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
5XR6
 
 | | Crystal structure of RabA1a in complex with GppNHp | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related protein RABA1a | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
1N4M
 
 | | Structure of Rb tumor suppressor bound to the transactivation domain of E2F-2 | | Descriptor: | Retinoblastoma Pocket, Transcription factor E2F2 | | Authors: | Lee, C, Chang, J.H, Lee, H.S, Cho, Y. | | Deposit date: | 2002-10-31 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
GENES DEV., 16, 2002
|
|
1N6A
 
 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N6C
 
 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
4V3I
 
 | | Crystal Structure of TssL from Vibrio cholerae. | | Descriptor: | GLYCEROL, VCA0115 | | Authors: | Jeong, J.H, Kim, Y.G. | | Deposit date: | 2014-10-19 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure of the bacterial type VI secretion system component TssL from Vibrio cholerae.
J. Microbiol., 53, 2015
|
|
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2G
 
 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2J
 
 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
