2J07
 
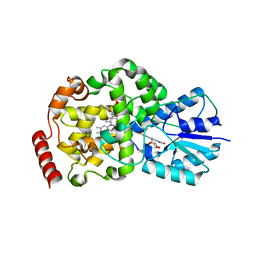 | | Thermus DNA photolyase with 8-HDF antenna chromophore | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J08
 
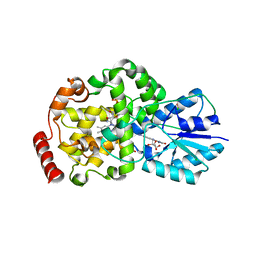 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
3CVV
 
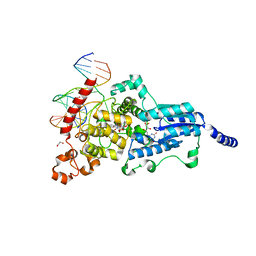 | | Drosophila melanogaster (6-4) photolyase bound to ds DNA with a T-T (6-4) photolesion and F0 cofactor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), ... | | Authors: | Glas, A.F, Maul, M.J, Cryle, M.J, Barends, T.R.M, Schneider, S, Kaya, E, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4RH0
 
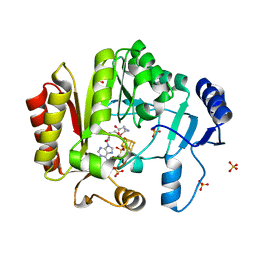 | | Spore photoproduct lyase C140A/S76C mutant with bound AdoMet | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Winkler, A, Carell, T, Schlichting, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rescuing DNA repair activity by rewiring the H-atom transfer pathway in the radical SAM enzyme, spore photoproduct lyase.
Chem.Commun.(Camb.), 50, 2014
|
|
4RH1
 
 | | Spore photoproduct lyase C140A/S76C mutant with bound AdoMet and dinucleoside spore photoproduct | | Descriptor: | 1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-[[(5R)-1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-methyl-2,4-bis(oxidanylidene)-1,3-diazinan-5-yl]methyl]pyrimidine-2,4-dione, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Winkler, A, Carell, T, Schlichting, I. | | Deposit date: | 2014-10-01 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rescuing DNA repair activity by rewiring the H-atom transfer pathway in the radical SAM enzyme, spore photoproduct lyase.
Chem.Commun.(Camb.), 50, 2014
|
|
1U45
 
 | | 8oxoguanine at the pre-insertion site of the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U4B
 
 | | Extension of an adenine-8oxoguanine mismatch | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U47
 
 | | cytosine-8-Oxoguanine base pair at the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U49
 
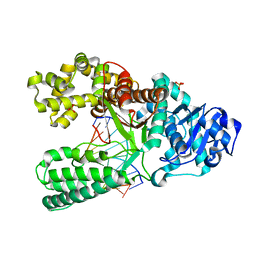 | | Adenine-8oxoguanine mismatch at the polymerase active site | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1U48
 
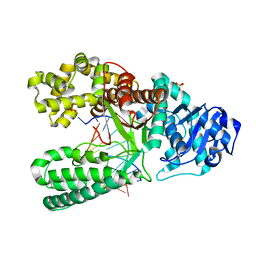 | | Extension of a cytosine-8-oxoguanine base pair | | Descriptor: | DNA polymerase I, DNA primer strand, DNA template strand with 8-oxoguanine, ... | | Authors: | Hsu, G.W, Ober, M, Carell, T, Beese, L.S. | | Deposit date: | 2004-07-23 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase.
Nature, 431, 2004
|
|
1TDZ
 
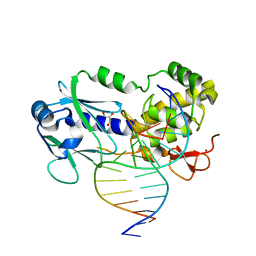 | | Crystal Structure Complex Between the Lactococcus Lactis FPG (Mutm) and a FAPY-dG Containing DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', GLYCEROL, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-05-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
2R7Z
 
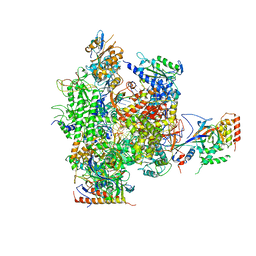 | | Cisplatin lesion containing RNA polymerase II elongation complex | | Descriptor: | 5'-D(*CP*AP*AP*GP*TP*AP*G)-3', 5'-D(*TP*AP*CP*TP*TP*GUP*CP*CP*CP*TP*CP*CP*TP*CP*AP*T)-3', 5'-R(*UP*UP*UP*GP*AP*GP*GP*AP*GP*G)-3', ... | | Authors: | Damsma, G.E, Alt, A, Brueckner, F, Carell, T, Cramer, P. | | Deposit date: | 2007-09-10 | | Release date: | 2007-11-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Mechanism of transcriptional stalling at cisplatin-damaged DNA.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2WQ7
 
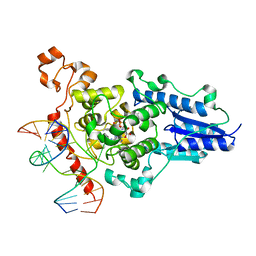 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(6-4)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*ZP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions.
J.Am.Chem.Soc., 132, 2010
|
|
2JA8
 
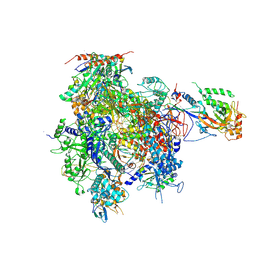 | | CPD lesion containing RNA Polymerase II elongation complex D | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TP*TTP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*AP*UP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2JA7
 
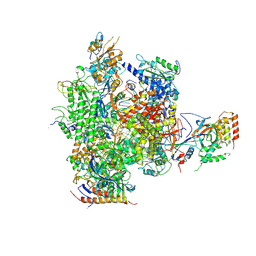 | | CPD lesion containing RNA Polymerase II elongation complex C | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TP*TTP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II.
Science, 315, 2007
|
|
2JA6
 
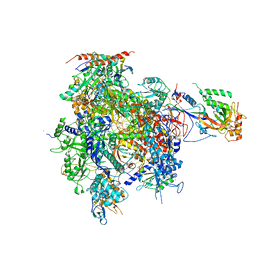 | | CPD lesion containing RNA Polymerase II elongation complex B | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TP*TTP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*TP*AP*AP*GP*TP*AP*CP*TP*TP*GP *AP*GP*CP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | CPD damage recognition by transcribing RNA polymerase II.
Science, 315, 2007
|
|
2JA5
 
 | | CPD lesion containing RNA Polymerase II elongation complex A | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*AP*GP*TP*AP *CP*TP*TTP*TP*TP*CP*CP*BRUP*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*AP)-3', DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Brueckner, F, Hennecke, U, Carell, T, Cramer, P. | | Deposit date: | 2006-11-23 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Cpd Damage Recognition by Transcribing RNA Polymerase II
Science, 315, 2007
|
|
2WQ6
 
 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(Dewar)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*CDWP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions
J.Am.Chem.Soc., 132, 2010
|
|
4B9M
 
 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dA DNA lesion -thymine basepair in the post- insertion site. | | Descriptor: | 5'-D(*DC*DA*DA*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4BW9
 
 | | PylRS Y306G, Y384F, I405R mutant in complex with AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schneider, S, Vrabel, M, Gattner, M.J, Fluegel, V, Lopez-Carillo, V, Carell, T. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Insights Into Incorporation of Norbornene Amino Acids for Click Modification of Proteins
Chem.Bio.Chem., 14, 2013
|
|
4B9U
 
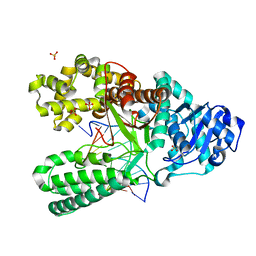 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dA basepair in the post-insertion site. | | Descriptor: | 5'-D(*CP*AP*AP*FOXP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4CIS
 
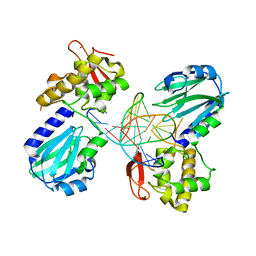 | | Structure of MutM in complex with carbocyclic 8-oxo-G containing DNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, DNA, FORMAMIDOPYRIMIDIN DNA GLYCOSYLASE, ... | | Authors: | Schneider, S, Sadeghian, K, Flaig, D, Blank, I.D, Strasser, R, Stathis, D, Winnacker, M, Carell, T, Ochsenfeld, C. | | Deposit date: | 2013-12-15 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ribose-protonated DNA base excision repair: a combined theoretical and experimental study.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4B9T
 
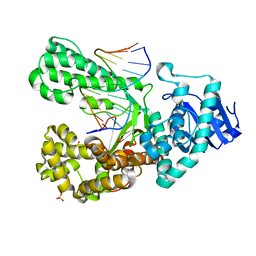 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion -dC basepair in the post-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9V
 
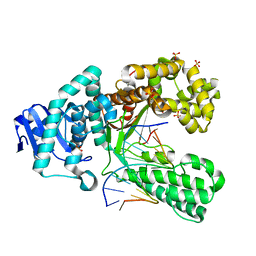 | | Structure of the high fidelity DNA polymerase I with extending from an oxidative formamidopyrimidine-dG DNA lesion -dA basepair. | | Descriptor: | 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*CP*CP*TP*GP*AP*CP*TP*CP*TP*AP*AP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4B9S
 
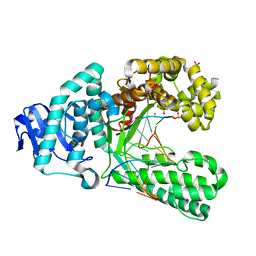 | | Structure of the high fidelity DNA polymerase I with an oxidative formamidopyrimidine-dG DNA lesion outside of the pre-insertion site. | | Descriptor: | 5'-D(*AP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', 5'-D(*CP*AP*TP*FOXP*AP*GP*AP*GP*TP*CP*AP*GP*GP*TP*TP)-3', DNA POLYMERASE, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-06 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
