8SIJ
 
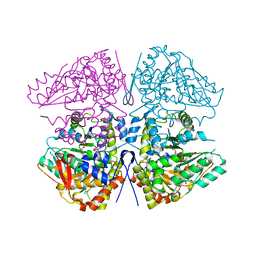 | | Crystal structure of F. varium tryptophanase | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Tryptophanase 1, ... | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
8SL7
 
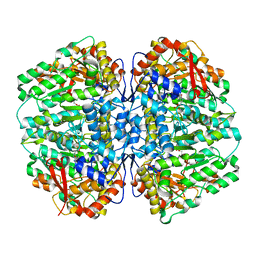 | | Butyricicoccus sp. BIOML-A1 tryptophanase complex with (3S) ALG-05 | | Descriptor: | (E)-3-[(3S)-3-chloro-2-oxo-2,3-dihydro-1H-indol-3-yl]-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, Tryptophanase | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
6CJ7
 
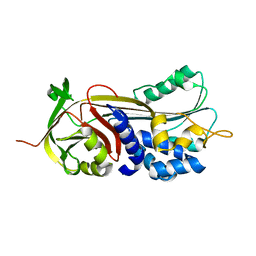 | | Crystal structure of Manduca sexta Serine protease inhibitor (Serpin)-12 | | Descriptor: | Serpin-12 | | Authors: | Gulati, M, Hu, Y, Peng, S, Pathak, P.K, Wang, Y, Deng, J, Jiang, H. | | Deposit date: | 2018-02-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Manduca sexta serpin-12 controls the prophenoloxidase activation system in larval hemolymph.
Insect Biochem. Mol. Biol., 99, 2018
|
|
6EE9
 
 | |
7CUN
 
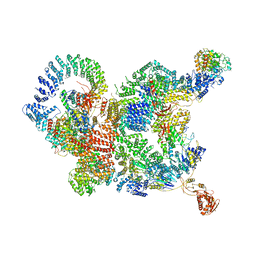 | | The structure of human Integrator-PP2A complex | | Descriptor: | Integrator complex subunit 1, Integrator complex subunit 11, Integrator complex subunit 2, ... | | Authors: | Zheng, H, Qi, Y, Liu, W, Li, J, Wang, J, Xu, Y. | | Deposit date: | 2020-08-23 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of Integrator-PP2A complex (INTAC), an RNA polymerase II phosphatase.
Science, 370, 2020
|
|
4RZH
 
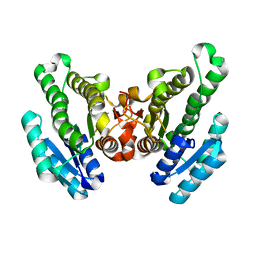 | | Crystal structure of FabG from Synechocystis sp. PCC 6803 | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase | | Authors: | Liu, Y, Xue, S. | | Deposit date: | 2014-12-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-directed construction of a high-performance version of the enzyme FabG from the photosynthetic microorganism Synechocystis sp. PCC 6803.
Febs Lett., 589, 2015
|
|
4RZI
 
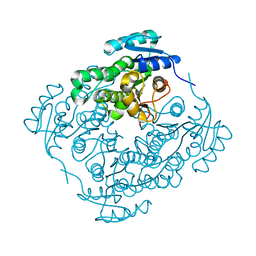 | | Crystal structure of PhaB from Synechocystis sp. PCC 6803 | | Descriptor: | 3-ketoacyl-acyl carrier protein reductase | | Authors: | Xue, S, Liu, Y. | | Deposit date: | 2014-12-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.891 Å) | | Cite: | Structure-directed construction of a high-performance version of the enzyme FabG from the photosynthetic microorganism Synechocystis sp. PCC 6803.
Febs Lett., 589, 2015
|
|
4RR5
 
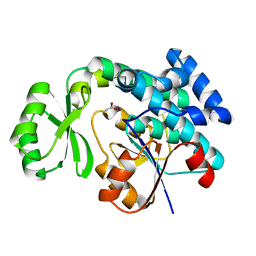 | | The crystal structure of Synechocystis sp. PCC 6803 Malonyl-CoA: ACP Transacylase | | Descriptor: | ACETATE ION, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Liu, Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | Structural and biochemical characterization of MCAT from photosynthetic microorganism Synechocystis sp. PCC 6803 reveal its stepwise catalytic mechanism
Biochem.Biophys.Res.Commun., 457, 2015
|
|
6KYF
 
 | | Crystal structure of an anti-CRISPR protein | | Descriptor: | AcrF11, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Niu, Y, Wang, H, Zhang, Y, Feng, Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A Type I-F Anti-CRISPR Protein Inhibits the CRISPR-Cas Surveillance Complex by ADP-Ribosylation.
Mol.Cell, 80, 2020
|
|
5TIW
 
 | |
5TA8
 
 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TA6
 
 | | Crystal structure of PLK1 in complex with a novel 5,6-dihydroimidazolo[1,5-f]pteridine inhibitor. | | Descriptor: | 4-{[(6R)-7-cyano-5-cyclopentyl-6-ethyl-5,6-dihydroimidazo[1,5-f]pteridin-3-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Serine/threonine-protein kinase PLK1, ZINC ION | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and SAR development of 5,6-dihydroimidazolo[1,5-f]pteridine derivatives as novel Polo-like kinase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TIX
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase/R-oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2R)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5TIV
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5TIY
 
 | | Schistosoma haematobium (Blood Fluke) Sulfotransferase/S-oxamniquine Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase, {(2S)-7-nitro-2-[(propan-2-ylamino)methyl]-1,2,3,4-tetrahydroquinolin-6-yl}methanol | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5TIZ
 
 | | Schistosoma japonicum (Blood Fluke) Sulfotransferase | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase | | Authors: | Taylor, A.B, Hart, P.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural and enzymatic insights into species-specific resistance to schistosome parasite drug therapy.
J. Biol. Chem., 292, 2017
|
|
5W54
 
 | |
3GQF
 
 | |
6KXG
 
 | | Crystal structure of caspase-11-CARD | | Descriptor: | caspase-11-CARD | | Authors: | Liu, M.Z.Y, Jin, T.C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Crystal structure of caspase-11 CARD provides insights into caspase-11 activation.
Cell Discov, 6, 2020
|
|
4J52
 
 | | Crystal structure of PLK1 in complex with a pyrimidodiazepinone inhibitor | | Descriptor: | 4-{[(7R)-9-cyclopentyl-7-ethenyl-7-fluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl]amino}-3-methoxy-N-(4-methylpiperazin-1-yl)benzamide, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Hosfield, D.J, Skene, R.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of TAK-960: An orally available small molecule inhibitor of polo-like kinase 1 (PLK1).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J53
 
 | | Crystal structure of PLK1 in complex with TAK-960 | | Descriptor: | 4-[(9-cyclopentyl-7,7-difluoro-5-methyl-6-oxo-6,7,8,9-tetrahydro-5H-pyrimido[4,5-b][1,4]diazepin-2-yl)amino]-2-fluoro-5-methoxy-N-(1-methylpiperidin-4-yl)benzamide, ACETATE ION, Serine/threonine-protein kinase PLK1, ... | | Authors: | Hosfield, D.J, Skene, R.J. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of TAK-960: An orally available small molecule inhibitor of polo-like kinase 1 (PLK1).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2KCJ
 
 | | solution structure of FAPP1 PH domain | | Descriptor: | Pleckstrin homology domain-containing family A member 3 | | Authors: | Lenoir, M, Coskun, U, James, J, Simons, K, Overduin, M. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-22 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis of wedging the Golgi membrane by FAPP pleckstrin homology domains.
Embo Rep., 11, 2010
|
|
2NNX
 
 | |
5YKO
 
 | |
5YKN
 
 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | Descriptor: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | Authors: | Yang, Z, Du, J. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
