1CEQ
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
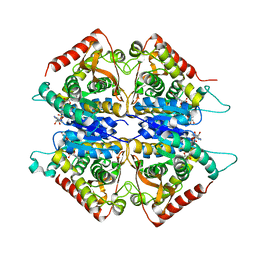
1CLO
 
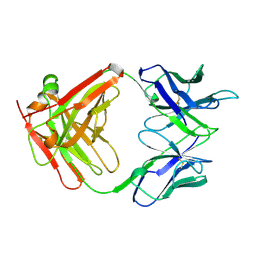 | |
2A5B
 
 | | Avidin complexed with 8-oxodeoxyguanosine | | Descriptor: | 2'-DEOXY-8-OXOGUANOSINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin | | Authors: | Conners, R, Hooley, E, Thomas, S, Brady, R.L. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Recognition of oxidatively modified bases within the biotin-binding site of avidin.
J.Mol.Biol., 357, 2006
|
|
4B7U
 
 | |
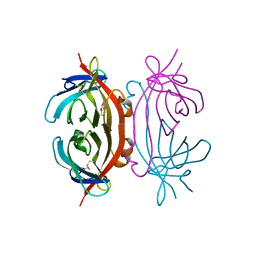
2A92
 
 | | Crystal structure of lactate dehydrogenase from Plasmodium vivax: complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
2A5C
 
 | | Structure of Avidin in complex with the ligand 8-oxodeoxyadenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-OXODEOXYADENOSINE, Avidin | | Authors: | Conners, R, Hooley, E, Thomas, S, Brady, R.L. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of oxidatively modified bases within the biotin-binding site of avidin.
J.Mol.Biol., 357, 2006
|
|
2A94
 
 | | Structure of Plasmodium falciparum lactate dehydrogenase complexed to APADH. | | Descriptor: | ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, L-lactate dehydrogenase | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-11 | | Release date: | 2006-01-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
2A8G
 
 | | Structure of Avidin in complex with the ligand deoxyguanosine | | Descriptor: | 2'-DEOXY-GUANOSINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin | | Authors: | Conners, R, Hooley, E, Thomas, S, Brady, R.L. | | Deposit date: | 2005-07-08 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Recognition of oxidatively modified bases within the biotin-binding site of avidin.
J.Mol.Biol., 357, 2006
|
|
2AA3
 
 | | Crystal structure of Plasmodium vivax lactate dehydrogenase complex with APADH | | Descriptor: | ACETYL PYRIDINE ADENINE DINUCLEOTIDE, REDUCED, L-lactate dehydrogenase, ... | | Authors: | Chaikuad, A, Fairweather, V, Conners, R, Joseph-Horne, T, Turgut-Balik, D, Brady, R.L. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Lactate Dehydrogenase from Plasmodium vivax: Complexes with NADH and APADH.
Biochemistry, 44, 2005
|
|
2AYG
 
 | |
2AYB
 
 | |
2AYE
 
 | |
4CUK
 
 | |
2CMY
 
 | | Crystal complex between bovine trypsin and Veronica hederifolia trypsin inhibitor | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, GLYCEROL, ... | | Authors: | Conners, R, Yardley, J.L, Konarev, A, Shewry, P, Brady, R.L. | | Deposit date: | 2006-05-16 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An Unusual Helix-Turn-Helix Protease Inhibitory Motif in a Novel Trypsin Inhibitor from Seeds of Veronica (Veronica Hederifolia L.).
J.Biol.Chem., 282, 2007
|
|
4CUJ
 
 | |
4DZN
 
 | | A de novo designed Coiled Coil CC-pIL | | Descriptor: | COILED-COIL PEPTIDE CC-PIL | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4DZM
 
 | | A de novo designed Coiled Coil CC-Di | | Descriptor: | COILED-COIL PEPTIDE CC-DI | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4DZK
 
 | | A de novo designed Coiled Coil CC-Tri-N13 | | Descriptor: | CHLORIDE ION, COILED-COIL PEPTIDE CC-TRI-N13 | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4DZL
 
 | | A de novo designed Coiled Coil CC-Tri | | Descriptor: | COILED-COIL PEPTIDE CC-TRI | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
6Q5N
 
 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-L24Nle | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CC-Hex*-L24Nle, GLYCEROL | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5H
 
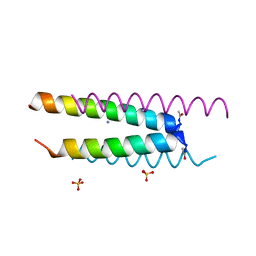 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24D | | Descriptor: | AMMONIUM ION, CC-Hex*-L24D, SULFATE ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5K
 
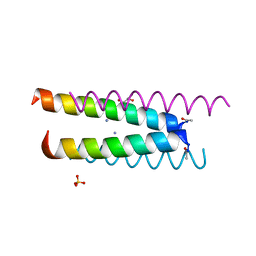 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24K | | Descriptor: | AMMONIUM ION, CC-Hex*-L24K, GLYCEROL, ... | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5Q
 
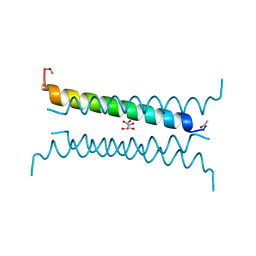 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex-KgEb | | Descriptor: | CC-Hex-KgEb, L(+)-TARTARIC ACID | | Authors: | Zaccai, N.R, Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5R
 
 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-LL-KgEb | | Descriptor: | CC-Hex*-LL-KgEb, GLYCEROL | | Authors: | Beesley, J.L, Rhys, G.G, Wood, C.W, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5J
 
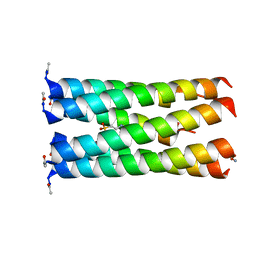 | | Crystal structure of a CC-Hex mutant that forms a parallel six-helix coiled coil CC-Hex*-L24E | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CC-Hex*-L24E | | Authors: | Rhys, G.G, Wood, C.W, Beesley, J.L, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
