4K89
 
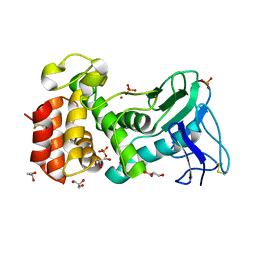 | | Crystal structure of Pseudomonas aeruginosa strain K solvent tolerant elastase | | Descriptor: | CALCIUM ION, GLYCEROL, Organic solvent tolerant elastase, ... | | Authors: | Ali, M.S.M, Said, Z.S.A.M, Rahman, R.N.Z.R.A, Basri, M, Salleh, A.B. | | Deposit date: | 2013-04-18 | | Release date: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structure analysia of solvent tolerant elastase strain K
To be Published
|
|
3UMJ
 
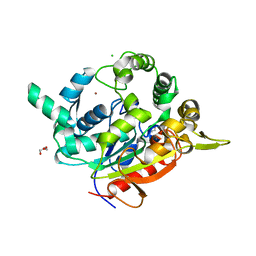 | | Crystal Structure of D311E Lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ruslan, R, Rahman, R.N.Z.R.A, Leow, T.C, Ali, M.S.M, Basri, M, Salleh, A.B. | | Deposit date: | 2011-11-13 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Thermal Stability via Outer-Loop Ion Pair Interaction of Mutated T1 Lipase from Geobacillus zalihae Strain T1
Int J Mol Sci, 13, 2012
|
|
4FDM
 
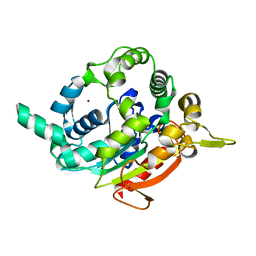 | | Crystallization and 3D structure elucidation of thermostable L2 lipase from thermophilic locally isolated Bacillus sp. L2. | | Descriptor: | CALCIUM ION, Thermostable lipase, ZINC ION | | Authors: | Rahman, R.N.Z.R.A, Shariff, F.M, Salleh, A.B, Basri, M.B. | | Deposit date: | 2012-05-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 3D Structure Elucidation of Thermostable L2 Lipase from Thermophilic Bacillus sp. L2.
Int.J.Mol.Sci., 13, 2012
|
|
2LQ1
 
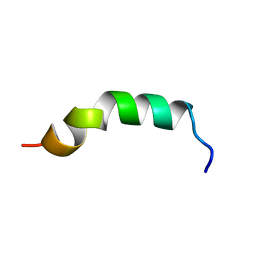 | | Solution structure of de novo designed antifreeze peptide 3 | | Descriptor: | de novo designed antifreeze peptide 3 | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2Z5G
 
 | | Crystal structure of T1 lipase F16L mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermostable lipase, ... | | Authors: | Matsumura, H, Yamamoto, T, Inoue, T, Kai, Y. | | Deposit date: | 2007-07-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
2DSN
 
 | | Crystal structure of T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Matsumura, H, Kai, Y. | | Deposit date: | 2006-07-03 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
2LQ0
 
 | | Solution structure of de novo designed antifreeze peptide 1m | | Descriptor: | de novo designed antifreeze peptide 1m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-21 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
2LQ2
 
 | | Solution structure of de novo designed peptide 4m | | Descriptor: | de novo designed antifreeze peptide 4m | | Authors: | Bhunia, A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures, dynamics, and ice growth inhibitory activity of Peptide fragments derived from an antarctic yeast protein
Plos One, 7, 2012
|
|
