5XG3
 
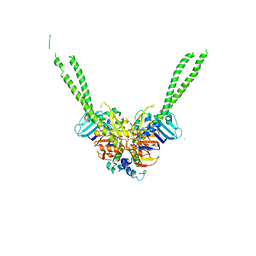 | | Crystal structure of the ATPgS-engaged Smc head domain with an extended coiled coil bound to the C-terminal domain of ScpA derived from Bacillus subtilis | | Descriptor: | COBALT (II) ION, Chromosome partition protein Smc, MAGNESIUM ION, ... | | Authors: | Shin, H.-C, Lee, H, Oh, B.-H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization
Mol. Cell, 67, 2017
|
|
5XG2
 
 | |
5XNS
 
 | |
3ZXI
 
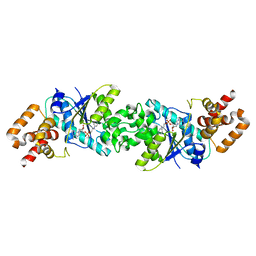 | | Crystal structure of human mitochondrial tyrosyl-tRNA synthetase in complex with a tyrosyl-adenylate analog | | Descriptor: | PHOSPHORIC ACID 2-AMINO-3-(4-HYDROXY-PHENYL)-PROPYL ESTER ADENOSIN-5'YL ESTER, TYROSYL-TRNA SYNTHETASE, MITOCHONDRIAL | | Authors: | Bonnefond, L, Frugier, M, Rudinger-Thirion, J, Balg, C, Chenevert, R, Lorber, B, Giege, R, Sauter, C. | | Deposit date: | 2011-08-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
2HSM
 
 | |
7OD1
 
 | |
7ONI
 
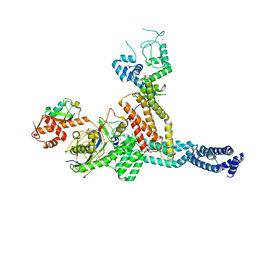 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | Descriptor: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | Authors: | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
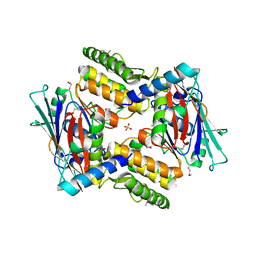
4I99
 
 | |
4I98
 
 | |
2HRA
 
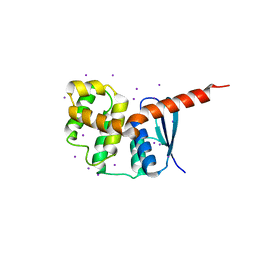 | | Crystal structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA aminoacylation and nuclear export cofactor Arc1p reveal a novel function for an old fold | | Descriptor: | Glutamyl-tRNA synthetase, cytoplasmic, IODIDE ION | | Authors: | Simader, H, Hothorn, M, Suck, D. | | Deposit date: | 2006-07-20 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the interacting domains from yeast glutamyl-tRNA synthetase and tRNA-aminoacylation and nuclear-export cofactor Arc1p reveal a novel function for an old fold.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
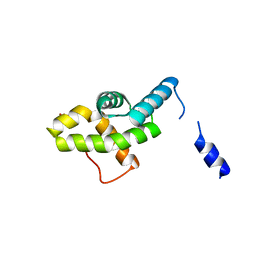
2HQT
 
 | |
