6K8Q
 
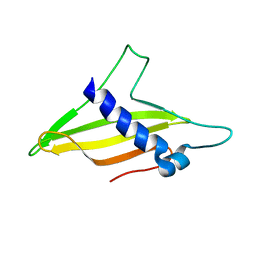 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6K7F
 
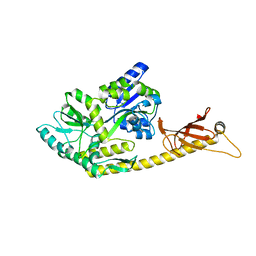 | | Crystal structure of MBPholo-Tim21 fusion protein with a 17-residue helical linker | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Mitochondrial import inner membrane translocase subunit TIM21, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bala, S, Shimada, A, Kohda, D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6K7D
 
 | |
6K7E
 
 | |
8VBZ
 
 | |
8VEQ
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with azlocillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-{[(2R)-2-{[(2-oxoimidazolidin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}ethyl]-1,3-thiazolidine-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
8VEN
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with cefoperazone | | Descriptor: | (2R,4R)-2-[(1R)-1-{[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-(4-hydroxyphenyl)acetyl]amino}-2-oxoethyl]-5-methylidene-1,3-thiazinane-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
8VEP
 
 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 acylated by piperacillin | | Descriptor: | DI(HYDROXYETHYL)ETHER, Piperacillin (Open Form), Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C.M, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
6LZB
 
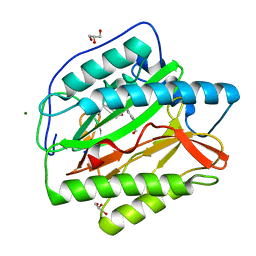 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with AN-P2-5H-06 | | Descriptor: | 1-[(3-methoxyphenyl)methyl]-~{N}-oxidanyl-pyrrolo[2,3-b]pyridine-5-carboxamide, COBALT (II) ION, GLYCEROL, ... | | Authors: | sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
6LZC
 
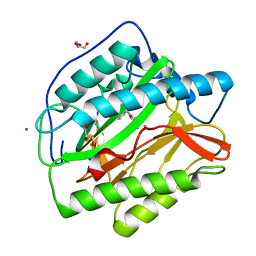 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with KV-P2-4H-05 | | Descriptor: | COBALT (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
8SKQ
 
 | |
6U7Y
 
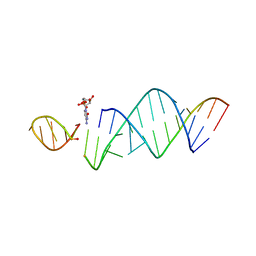 | | RNA hairpin structure containing one TNA nucleotide as template | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*(TG)P*CP*CP*GP*AP*AP*AP*GP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
5H5A
 
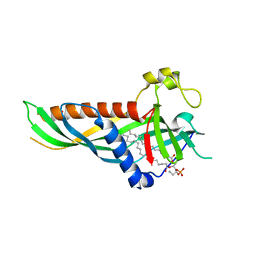 | | Mdm12 from K. lactis (1-239), Lys residues are uniformly dimethyl modified | | Descriptor: | Mitochondrial distribution and morphology protein 12, POTASSIUM ION, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H54
 
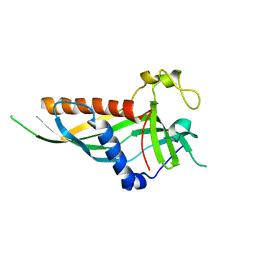 | | Mdm12 from K. lactis 1-239 | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H55
 
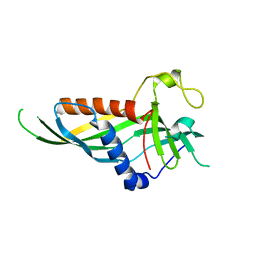 | | Mdm12 from K. lactis | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H5C
 
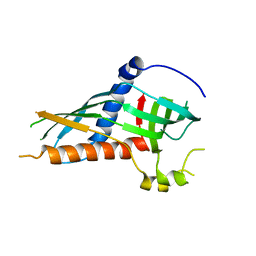 | |
7DVI
 
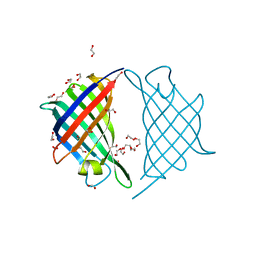 | |
7DVK
 
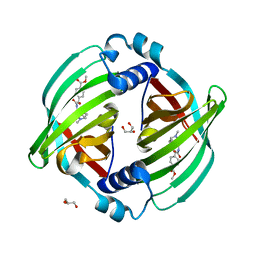 | | PyrI4 in complex with intermolecular Diels-Alder product | | Descriptor: | GLYCEROL, Spiro-conjugate synthase, methyl 4-[[(1S,6S)-6-(dimethylcarbamoyl)cyclohex-2-en-1-yl]carbamoyloxymethyl]benzoate | | Authors: | Kashyap, R, Addlagatta, A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exo-selective intermolecular Diels-Alder reaction by PyrI4 and AbnU on non-natural substrates.
Commun Chem, 2021
|
|
6U7Z
 
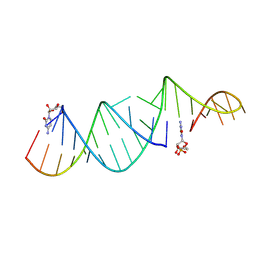 | | RNA hairpin structure containing one TNA nucleotide as primer | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*GP*CP*CP*GP*AP*AP*AP*GP*(TG))-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6U8U
 
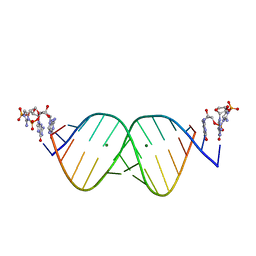 | | RNA duplex bound with TNA 3'-3' imidazolium dimer | | Descriptor: | 2-amino-3-[(R)-{[(3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-hydroxyoxolan-3-yl]oxy}(hydroxy)phosphoryl]-1-[(S)-{[(3S,4R,5R)-5-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-4-hydroxyoxolan-3-yl]oxy}(hydroxy)phosphoryl]-1H-imidazol-3-ium, MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-05 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6U89
 
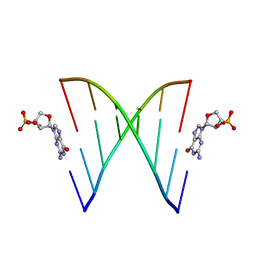 | | RNA duplex, bound with TNA monomer | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6U8F
 
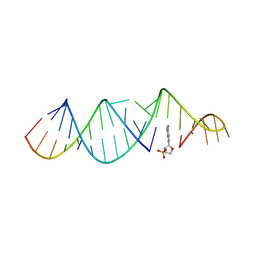 | | RNA hairpin, bound with TNA monomer | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*AP*UP*GP*AP*AP*AP*GP*U)-3'), RNA (5'-R(P*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
