7E4L
 
 | |
5HN4
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and homoisocitrate | | Descriptor: | (1R,2S)-1-hydroxybutane-1,2,4-tricarboxylic acid, Homoisocitrate dehydrogenase, IMIDAZOLE, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN6
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and 3-isopropylmalate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-ISOPROPYLMALIC ACID, Homoisocitrate dehydrogenase, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HN5
 
 | | Crystal structure of beta-decarboxylating dehydrogenase (TK0280) from Thermococcus kodakarensis complexed with Mn and isocitrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Homoisocitrate dehydrogenase, ISOCITRIC ACID, ... | | Authors: | Shimizu, T, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-01-18 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and function of an ancestral-type beta-decarboxylating dehydrogenase from Thermococcus kodakarensis
Biochem. J., 474, 2017
|
|
5HWS
 
 | |
5HN3
 
 | |
5AYV
 
 | | Crystal structure of archaeal ketopantoate reductase complexed with coenzyme A and 2-oxopantoate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-dehydropantoate 2-reductase, ACETATE ION, ... | | Authors: | Aikawa, Y, Nishitani, Y, Miki, K. | | Deposit date: | 2015-09-08 | | Release date: | 2016-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of archaeal ketopantoate reductase complexed with coenzyme a and 2-oxopantoate provides structural insights into feedback regulation
Proteins, 84, 2016
|
|
2YH2
 
 | | Pyrobaculum calidifontis esterase monoclinic form | | Descriptor: | ESTERASE, SULFATE ION | | Authors: | Palm, G.J, Bogdanovic, X, Hinrichs, W. | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of an Esterase Fom the Hyperthermophilic Microorganism Pyrobaculum Calidifontis Va1 Supports Explanation of its Enantioselectivity.
Appl.Microbiol.Biotechnol., 91, 2011
|
|
5HEE
 
 | | Crystal structure of the TK2203 protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5IJA
 
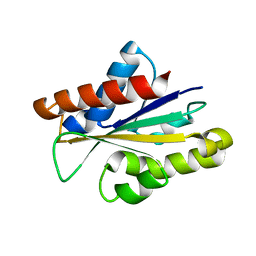 | |
3ZWQ
 
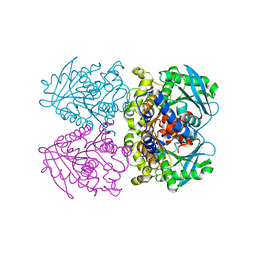 | |
5AUQ
 
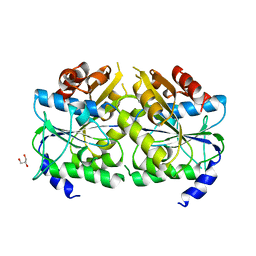 | | Crystal structure of ATPase-type HypB in the nucleotide free state | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUP
 
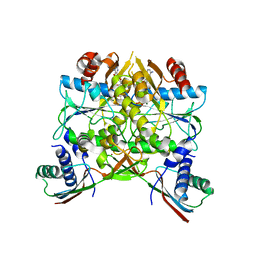 | | Crystal structure of the HypAB complex | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUN
 
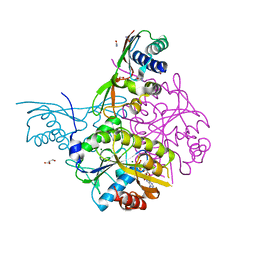 | | Crystal structure of the HypAB-Ni complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase involved in chromosome partitioning, ParA/MinD family, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5AUO
 
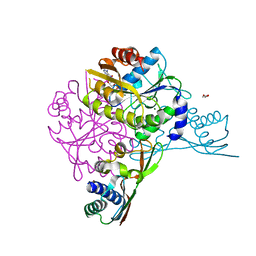 | | Crystal structure of the HypAB-Ni complex (AMPPCP) | | Descriptor: | ATPase involved in chromosome partitioning, ParA/MinD family, Mrp homolog, ... | | Authors: | Watanabe, S, Kawashima, T, Nishitani, Y, Miki, K. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DHE
 
 | | Crystal structure of ChBD3 from Thermococcus kodakarensis KOD1 | | Descriptor: | Chitinase, GLYCEROL | | Authors: | Niwa, S, Hibi, M, Takeda, K, Miki, K. | | Deposit date: | 2015-08-30 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of chitin binding domains of chitinase from Thermococcus kodakarensis KOD1
Febs Lett., 590, 2016
|
|
5DHD
 
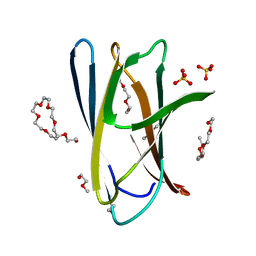 | | Crystal structure of ChBD2 from Thermococcus kodakarensis KOD1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Chitinase, SULFATE ION | | Authors: | Hibi, M, Niwa, S, Takeda, K, Miki, K. | | Deposit date: | 2015-08-30 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structures of chitin binding domains of chitinase from Thermococcus kodakarensis KOD1
Febs Lett., 590, 2016
|
|
3A43
 
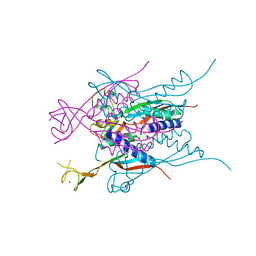 | | Crystal structure of HypA | | Descriptor: | Hydrogenase nickel incorporation protein hypA, ZINC ION | | Authors: | Watanabe, S, Arai, T, Matsumi, R, Aromi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-06-30 | | Release date: | 2009-10-06 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation.
J.Mol.Biol., 394, 2009
|
|
5X0G
 
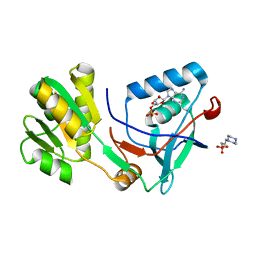 | | Free serine kinase (E30A mutant) in complex with ADP | | Descriptor: | 2-hydroxy-3-[4-(2-hydroxy-3-sulfopropyl)piperazin-1-yl]propane-1-sulfonic acid, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
5X0J
 
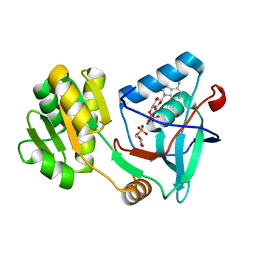 | | Free serine kinase (E30Q mutant) in complex with phosphoserine and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Free serine kinase, MAGNESIUM ION, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
5X0F
 
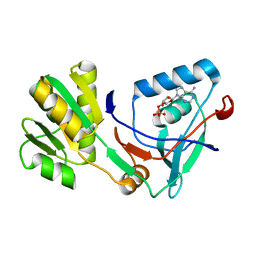 | |
5X0K
 
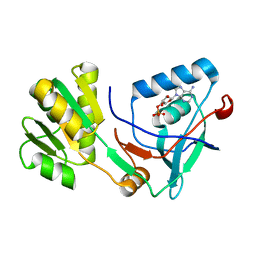 | |
5X0B
 
 | | Free serine kinase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Free serine kinase | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
5X0E
 
 | | Free serine kinase (E30A mutant) in complex with phosphoserine and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Free serine kinase, MAGNESIUM ION, ... | | Authors: | Nagata, R, Fujihashi, M, Miki, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Study on the Reaction Mechanism of a Free Serine Kinase Involved in Cysteine Biosynthesis
ACS Chem. Biol., 12, 2017
|
|
3W4S
 
 | |
