2C21
 
 | | Specificity of the Trypanothione-dependednt Leishmania major Glyoxalase I: Structure and biochemical comparison with the human enzyme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, NICKEL (II) ION, ... | | Authors: | Ariza, A, Vickers, T.J, Greig, N, Armour, K.A, Eggleston, I.M, Fairlamb, A.H, Bond, C.S. | | Deposit date: | 2005-09-23 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of the Trypanothione-Dependent Leishmania Major Glyoxalase I: Structure and Biochemical Comparison with the Human Enzyme.
Mol.Microbiol., 59, 2006
|
|
2BKE
 
 | | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal RadA | | Descriptor: | CHLORIDE ION, DNA REPAIR AND RECOMBINATION PROTEIN RADA | | Authors: | Ariza, A, Richard, D.L, White, M.F, Bond, C.S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal Rada
Nucleic Acids Res., 33, 2005
|
|
3ZQ9
 
 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | (2R,3S,4R,5R)-5-(HYDROXYMETHYL)PIPERIDINE-2,3,4-TRIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
3ZLA
 
 | | Crystal structure of the nucleocapsid protein from Bunyamwera virus bound to RNA | | Descriptor: | NUCLEOPROTEIN, RNA | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
3ZL9
 
 | | Crystal structure of the nucleocapsid protein from Schmallenberg virus | | Descriptor: | NUCLEOCAPSID PROTEIN | | Authors: | Ariza, A, Tanner, S.J, Walter, C.T, Dent, K.C, Shepherd, D.A, Wu, W, Matthews, S.V, Hiscox, J.A, Green, T.J, Luo, M, Elliot, R.M, Ashcroft, A.E, Stonehouse, N.J, Ranson, N.A, Barr, J.N, Edwards, T.A. | | Deposit date: | 2013-01-29 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Nucleocapsid Protein Structures from Orthobunyaviruses Reveal Insight Into Ribonucleoprotein Architecture and RNA Polymerization.
Nucleic Acids Res., 41, 2013
|
|
2YIH
 
 | | Structure of a Paenibacillus polymyxa Xyloglucanase from GH family 44 with Xyloglucan | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-13 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
2YKK
 
 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
2YJQ
 
 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-23 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
4ARS
 
 | | Hafnia Alvei phytase apo form | | Descriptor: | ACETATE ION, GLYCEROL, HISTIDINE ACID PHOSPHATASE | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
4ARU
 
 | | Hafnia Alvei phytase in complex with tartrate | | Descriptor: | CHLORIDE ION, HISTIDINE ACID PHOSPHATASE, L(+)-TARTARIC ACID, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
4ARV
 
 | | Yersinia kristensenii phytase apo form | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
8COB
 
 | |
7AKR
 
 | |
7AKS
 
 | |
5LW0
 
 | | Oryza sativa APL macrodomain in complex with ADP-ribose | | Descriptor: | Basic helix-loop-helix, putative, expressed, ... | | Authors: | Ariza, A. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The role of ADP-ribosylation in regulating DNA interstrand crosslink repair.
J.Cell.Sci., 129, 2016
|
|
5M3I
 
 | | Macrodomain of Mycobacterium tuberculosis DarG | | Descriptor: | CHLORIDE ION, RNase III inhibitor | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5M3E
 
 | | Macrodomain of Thermus aquaticus DarG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Appr-1-p processing domain protein, CHLORIDE ION | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5M31
 
 | | Macrodomain of Thermus aquaticus DarG | | Descriptor: | Appr-1-p processing domain protein, CHLORIDE ION, GLYCEROL | | Authors: | Ariza, A. | | Deposit date: | 2016-10-13 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
5MKW
 
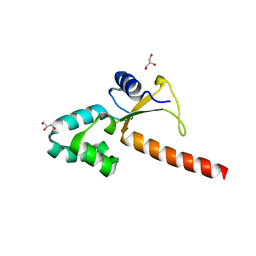 | | Crystal structure of the human ZRANB3 HNH domain | | Descriptor: | DNA annealing helicase and endonuclease ZRANB3, GLYCEROL, ZINC ION | | Authors: | Ariza, A. | | Deposit date: | 2016-12-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the function of ZRANB3 in replication stress response.
Nat Commun, 8, 2017
|
|
5MLO
 
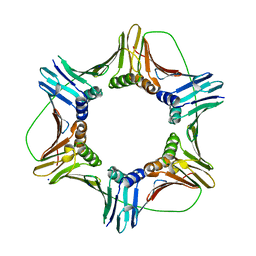 | |
5MLW
 
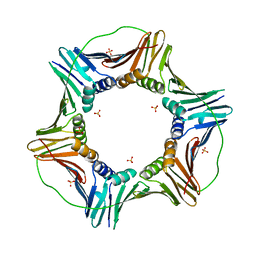 | |
7OMU
 
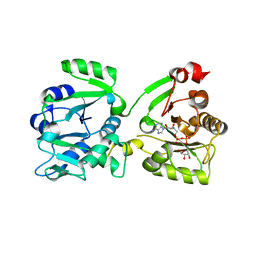 | | Thermosipho africanus DarTG in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Macro domain-containing protein | | Authors: | Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
8ADK
 
 | |
8ADJ
 
 | | Poly(ADP-ribose) glycohydrolase (PARG) from Drosophila melanogaster in complex with PARG inhibitor PDD00017272 | | Descriptor: | 1-[(2,5-dimethylpyrazol-3-yl)methyl]-N-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ariza, A, Fontana, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling.
Nat Commun, 14, 2023
|
|
6TVH
 
 | |
