7KW6
 
 | | Crystal structure of the BlCel48B from Bacillus licheniformis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exoglucanase-2, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2020-11-30 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Impact of cellulose properties on enzymatic degradation by bacterial GH48 enzymes: Structural and mechanistic insights from processive Bacillus licheniformis Cel48B cellulase.
Carbohydr Polym, 264, 2021
|
|
8U4A
 
 | |
8U4F
 
 | |
8U49
 
 | | The Apo Crystal Structure of BlCel9A from Glycoside Hydrolase Family 9 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-09-10 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of cellulose depolymerization by the two-domain BlCel9A enzyme from the glycoside hydrolase family 9.
Carbohydr Polym, 329, 2024
|
|
7KN8
 
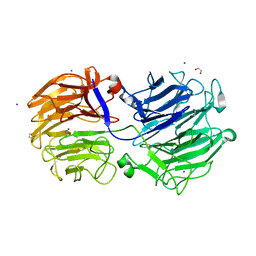 | | Crystal structure of the GH74 xyloglucanase from Xanthomonas campestris (Xcc1752) | | Descriptor: | 1,2-ETHANEDIOL, Cellulase, IODIDE ION, ... | | Authors: | Araujo, E.A, Vieira, P.S, Murakami, M.T, Polikarpov, I. | | Deposit date: | 2020-11-04 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Xyloglucan processing machinery in Xanthomonas pathogens and its role in the transcriptional activation of virulence factors
Nature Communications, 12, 2021
|
|
6WQW
 
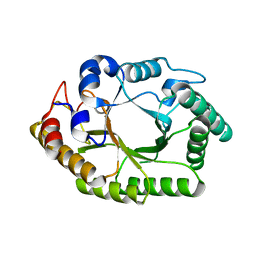 | | Thermobacillus composti GH10 xylanase | | Descriptor: | Beta-xylanase, beta-D-xylopyranose | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-04-29 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Transformation of xylan into value-added biocommodities using Thermobacillus composti GH10 xylanase.
Carbohydr Polym, 247, 2020
|
|
7KMN
 
 | |
7KMP
 
 | |
7KMM
 
 | |
7KMO
 
 | |
7KMQ
 
 | |
7KNC
 
 | |
8GF3
 
 | | Crystallographic structure from BlMan5_7 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Briganti, L, Araujo, E.A, Polikarpov, I. | | Deposit date: | 2023-03-07 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic structure from BlMan5_7
To be published
|
|
6O9N
 
 | | Structural insights on a new fungal aryl-alcohol oxidase | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kadowaki, M.A.S, Polikarpov, I. | | Deposit date: | 2019-03-14 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Enzymatic versatility and thermostability of a new aryl-alcohol oxidase from Thermothelomyces thermophilus M77.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
4YZT
 
 | | Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose | | Descriptor: | 1,2-ETHANEDIOL, Cellulose hydrolase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Popov, A, Polikarpov, I. | | Deposit date: | 2015-03-25 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism.
Sci Rep, 6, 2016
|
|
4YZP
 
 | |
