6WYB
 
 | | RTX (Reverse Transcription Xenopolymerase) in complex with an RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, DNA polymerase, ... | | Authors: | Choi, W.S, He, P, Pothukuchy, A, Gollihar, J, Ellington, A.D, Yang, W. | | Deposit date: | 2020-05-12 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a B family DNA polymerase has been evolved to copy RNA.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8OYF
 
 | |
8OYG
 
 | |
8P91
 
 | |
8OVB
 
 | |
8OSQ
 
 | |
8P9M
 
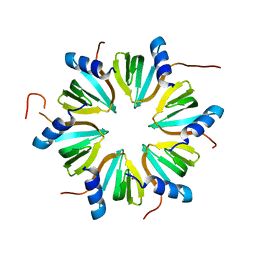 | | Hexameric Hfq from Chromobacterium haemolyticum | | Descriptor: | GLYCEROL, RNA-binding protein Hfq | | Authors: | Nikulin, A.D, Lekontseva, N.V. | | Deposit date: | 2023-06-06 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Hfq Protein from Chromobacterium haemolyticum Revealed a New Variant of Regulation of RNA Binding with the Protein
Crystallography Reports, 68, 2023
|
|
8QAQ
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. Conformation 1 of omphalotin A in apolar solvents. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QAS
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-23 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8Q7J
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability | | Descriptor: | CYCLOSPORIN A | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QBP
 
 | | Conformations of macrocyclic peptides sampled by exact NOEs: models for cell-permeability. NMR structure of Omphalotin A in methanol / water indoleOut conformation. | | Descriptor: | TRP-MVA-ILE-MVA-MVA-SAR-MVA-IML-SAR-VAL-IML-SAR | | Authors: | Ruedisser, S.H, Matabaro, E, Sonderegger, L, Guentert, P, Kuenzler, M, Gossert, A.D. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | SOLUTION NMR | | Cite: | Conformations of Macrocyclic Peptides Sampled by Nuclear Magnetic Resonance: Models for Cell-Permeability.
J.Am.Chem.Soc., 145, 2023
|
|
8QSO
 
 | | Crystal structure of human Mcl-1 in complex with compound 1 | | Descriptor: | (13S,16R,19S)-16-benzyl-43-ethoxy-N-methyl-7,11,14,17-tetraoxo-13-phenyl-5-oxa-2,8,12,15,18-pentaaza-1(1,4),4(1,2)-dibenzena-9(1,4)-cyclohexanacycloicosaphane-19-carboxamide, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hekking, K.F.W, Gremmen, S, Maroto, S, Keefe, A.D, Zhang, Y. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Development of Potent Mcl-1 Inhibitors: Structural Investigations on Macrocycles Originating from a DNA-Encoded Chemical Library Screen.
J.Med.Chem., 67, 2024
|
|
8SPF
 
 | | Crystal structure of Bax core domain BH3-groove dimer - hexameric fraction with 2-stearoyl lysoPC | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DODECANE, ... | | Authors: | Cowan, A.D, Miller, M.S, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 2023
|
|
8SPZ
 
 | |
8SPE
 
 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P31 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 2023
|
|
8SLS
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and ambient pressure (0.1 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLU
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and high pressure (205 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLT
 
 | | Crystal structure of human STEP (PTPN5) at physiological temperature (310 K) and ambient pressure (0.1 MPa) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SVT
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-331 | | Descriptor: | DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1, ... | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR
Nat Commun, 2024
|
|
8SZV
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog SJPYT-318 | | Descriptor: | N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-[(4-phenoxyphenyl)methyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Li, Y, Chen, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand flexibility and binding pocket malleability cooperate to allow selective PXR activation by analogs of a promiscuous nuclear receptor ligand.
Structure, 31, 2023
|
|
6O5Z
 
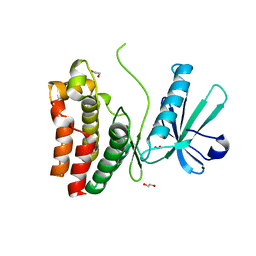 | | Crystal Structure of the human MLKL pseudokinase domain bound to compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-fluoranyl-5-(trifluoromethyl)phenyl]-3-[4-[methyl-[2-[(3-sulfamoylphenyl)amino]pyrimidin-4-yl]amino]phenyl]urea, Mixed lineage kinase domain-like protein | | Authors: | Cowan, A.D, Murphy, J.M, Pierotti, C.L, Lessene, G.L, Czabotar, P.E. | | Deposit date: | 2019-03-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Potent Inhibition of Necroptosis by Simultaneously Targeting Multiple Effectors of the Pathway.
Acs Chem.Biol., 15, 2020
|
|
6Q39
 
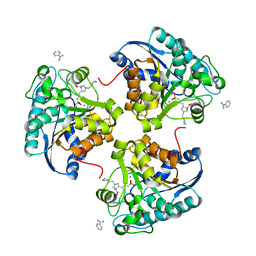 | | Complex of Arginase 2 with Example 49 | | Descriptor: | 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-1-[[(2~{S})-piperidin-2-yl]methyl]pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, mitochondrial, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.213 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
6Q37
 
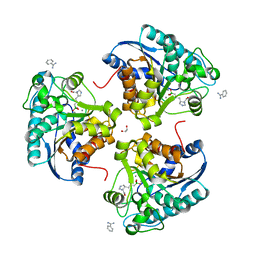 | | Complex of Arginase 2 with Example 23 | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3~{S},4~{R})-4-azanyl-4-carboxy-pyrrolidin-3-yl]propyl-tris(oxidanyl)boranuide, Arginase-2, ... | | Authors: | Podjarny, A.D, Van Zandt, M.C, Cousido-Siah, A. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Discovery ofN-Substituted 3-Amino-4-(3-boronopropyl)pyrrolidine-3-carboxylic Acids as Highly Potent Third-Generation Inhibitors of Human Arginase I and II.
J.Med.Chem., 62, 2019
|
|
6R1B
 
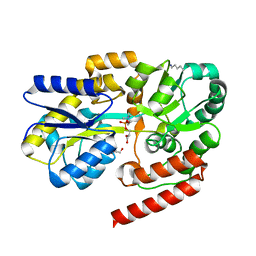 | | Crystal structure of UgpB from Mycobacterium tuberculosis in complex with glycerophosphocholine | | Descriptor: | 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fenn, J, Nepravishta, R, Guy, C.S, Harrison, J, Angulo, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27000213 Å) | | Cite: | Structural Basis of Glycerophosphodiester Recognition by theMycobacterium tuberculosisSubstrate-Binding Protein UgpB.
Acs Chem.Biol., 14, 2019
|
|
6RVV
 
 | | Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
