1T33
 
 | |
5D68
 
 | | Crystal structure of KRIT1 ARD-FERM | | Descriptor: | Krev interaction trapped protein 1 | | Authors: | Zhang, R, Li, X, Boggon, T.J. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural analysis of the KRIT1 ankyrin repeat and FERM domains reveals a conformationally stable ARD-FERM interface.
J.Struct.Biol., 192, 2015
|
|
8U1T
 
 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Zhang, R, Qin, H, Prasad, R, Fu, R, Zhou, H.X, Cross, T. | | Deposit date: | 2023-09-02 | | Release date: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein.
Commun Biol, 6, 2023
|
|
8JIF
 
 | | Cryo-EM Structure of 3-axis block of AAV9P31-Car4 complex | | Descriptor: | Capsid protein VP1, Carbonic anhydrase 4, ZINC ION | | Authors: | Zhang, R, Liu, Y, Lou, Z. | | Deposit date: | 2023-05-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Structural basis of the recognition of adeno-associated virus by the neurological system-related receptor carbonic anhydrase IV.
Plos Pathog., 20, 2024
|
|
3GOS
 
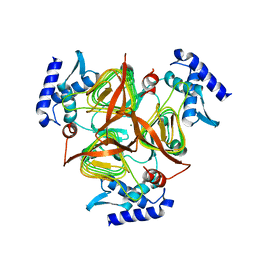 | | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Yersinia pestis CO92 | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, MAGNESIUM ION | | Authors: | Zhang, R, Maltseva, N, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-19 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Yersinia pestis CO92
To be Published
|
|
3GLV
 
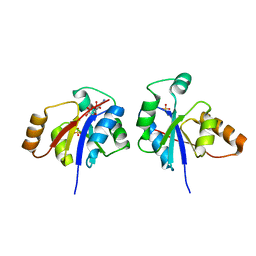 | | Crystal structure of the lipopolysaccharide core biosynthesis protein from Thermoplasma volcanium GSS1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Lipopolysaccharide core biosynthesis protein, SULFATE ION | | Authors: | Zhang, R, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the lipopolysaccharide core biosynthesis protein from Thermoplasma volcanium GSS1
To be Published
|
|
3H04
 
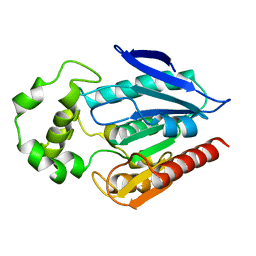 | | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | uncharacterized protein | | Authors: | Zhang, R, Tesar, C, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-08 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of the protein with unknown function from Staphylococcus aureus subsp. aureus Mu50
To be Published
|
|
3HCY
 
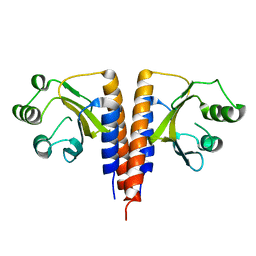 | | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021 | | Descriptor: | Putative two-component sensor histidine kinase protein | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021
To be Published
|
|
3HID
 
 | | Crystal structure of adenylosuccinate synthetase from Yersinia pestis CO92 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Adenylosuccinate synthetase | | Authors: | Zhang, R, Zhou, M, Peterson, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-19 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the adenylosuccinate synthetase from Yersinia pestis CO92
To be Published
|
|
3HFI
 
 | | The crystal structure of the putative regulator from Escherichia coli CFT073 | | Descriptor: | Putative regulator | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-11 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the putative regulator from Escherichia coli CFT073
To be Published
|
|
3IPJ
 
 | | The crystal structure of one domain of the PTS system, IIabc component from Clostridium difficile | | Descriptor: | PTS system, IIabc component, ZINC ION | | Authors: | Zhang, R, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-17 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of one domain of the PTS system, IIabc component from Clostridium difficile
To be Published
|
|
3IL0
 
 | | The crystal structure of the aminopeptidase P,XAA-pro aminopeptidase from Streptococcus thermophilus | | Descriptor: | Aminopeptidase P; XAA-pro aminopeptidase, GLYCEROL | | Authors: | Zhang, R, Hatzos, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the aminopeptidase P,XAA-pro aminopeptidase from Streptococcus thermophilus
To be Published
|
|
3I99
 
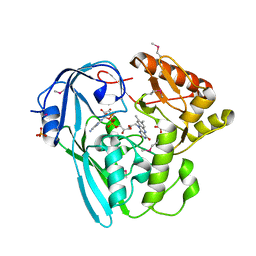 | | The crystal structure of the UDP-N-acetylenolpyruvoylglucosamine reductase from the Vibrio cholerae O1 biovar Tor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Zhang, R, Gu, M, Peterson, S, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the UDP-N-acetylenolpyruvoylglucosamine reductase from the Vibrio cholerae O1 biovar Tor
To be Published
|
|
3IC4
 
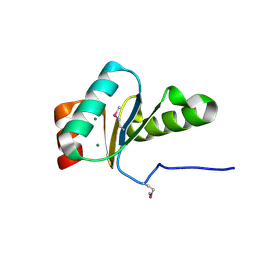 | | The crystal structure of the glutaredoxin(grx-1) from Archaeoglobus fulgidus | | Descriptor: | Glutaredoxin (Grx-1), MAGNESIUM ION | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the glutaredoxin from Archaeoglobus fulgidus
To be Published
|
|
3JAT
 
 | |
3JAS
 
 | |
3JAR
 
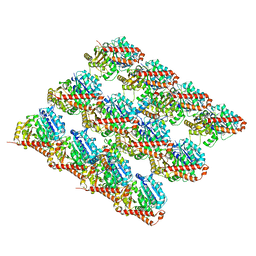 | | Cryo-EM structure of GDP-microtubule co-polymerized with EB3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2015-06-19 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins.
Cell(Cambridge,Mass.), 162, 2015
|
|
3JAW
 
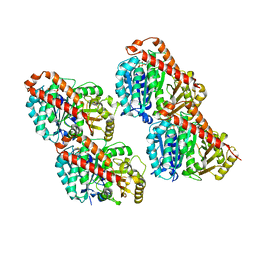 | | Atomic model of a microtubule seam based on a cryo-EM reconstruction of the EB3-bound microtubule (merged dataset containing tubulin bound to GTPgammaS, GMPCPP, and GDP) | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2015-06-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins.
Cell(Cambridge,Mass.), 162, 2015
|
|
3I12
 
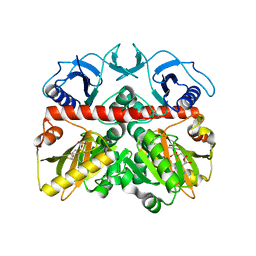 | | The crystal structure of the D-alanyl-alanine synthetase A from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase A | | Authors: | Zhang, R, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the D-alanyl-alanine synthetase A from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
3JAL
 
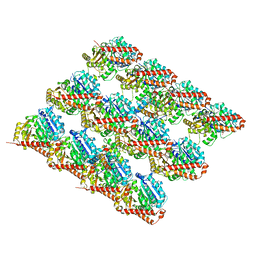 | | Cryo-EM structure of GMPCPP-microtubule co-polymerized with EB3 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Microtubule-associated protein RP/EB family member 3, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2015-06-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins.
Cell(Cambridge,Mass.), 162, 2015
|
|
3JAK
 
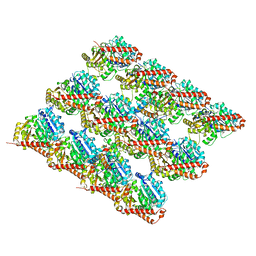 | |
3JR7
 
 | | The crystal structure of the protein of DegV family COG1307 with unknown function from Ruminococcus gnavus ATCC 29149 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the protein of DegV family COG1307 with unknown function from Ruminococcus gnavus ATCC 29149
To be Published
|
|
3GMI
 
 | |
3H92
 
 | |
3KBQ
 
 | |
