6E0C
 
 | |
6DZT
 
 | |
7RE9
 
 | | TCR mimic antibody (Fab fragment) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab heavy chain, Fab light chain | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
7RE8
 
 | |
7RE7
 
 | | TCR mimic antibody (Fab fragment) in complex with AFP/HLA-A*02 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, FORMIC ACID, ... | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
1B4P
 
 | | CRYSTAL STRUCTURES OF CLASS MU CHIMERIC GST ISOENZYMES M1-2 AND M2-1 | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, PROTEIN (GLUTATHIONE S-TRANSFERASE), SULFATE ION | | Authors: | Xiao, G, Chen, J, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1998-12-26 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Class MU Chimeric GST Isoenzymes M1-2 and M2-1
To be Published
|
|
6GSY
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3 | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
6GSV
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
1DK9
 
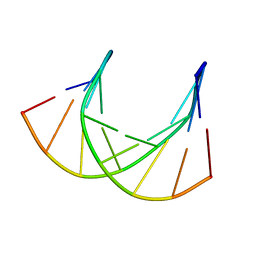 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 1999-12-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1DK6
 
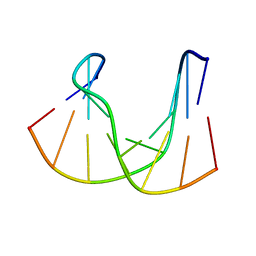 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
3NTE
 
 | |
3GST
 
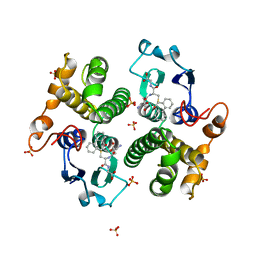 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
6OLU
 
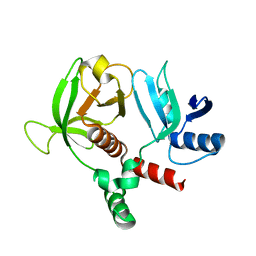 | | RIAM RA-PH core structure in the P212121 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation of RIAM by src promotes integrin activation by unmasking the PH domain of RIAM.
Structure, 29, 2021
|
|
6O6H
 
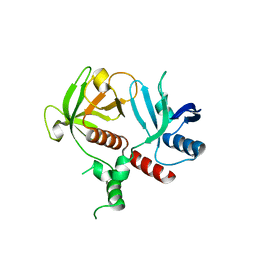 | | RIAM cc-RA-PH structure in the P21212 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation of RIAM by Src Promotes Integrin Activation by Unmasking the PH Domain of RIAM.
Structure, 2020
|
|
7SSM
 
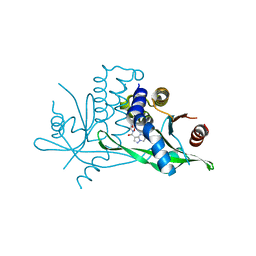 | | Crystal structure of human STING R232 in complex with compound 11 | | Descriptor: | 2-({[(8R)-pyrazolo[1,5-a]pyrimidine-3-carbonyl]amino}methyl)-1-benzofuran-7-carboxylic acid, Stimulator of interferon genes protein | | Authors: | Sack, J.S, Critton, D.A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Non-Nucleotide Small-Molecule STING Agonists via Chemotype Hybridization.
J.Med.Chem., 65, 2022
|
|
6SB1
 
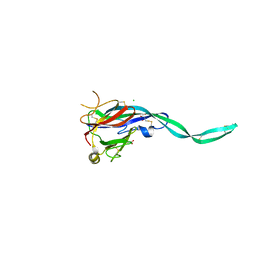 | | Crystal structure of murine perforin-2 P2 domain crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
6SB3
 
 | |
6SB4
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 2 | | Descriptor: | Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Yu, X, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
6SB5
 
 | |
7MJU
 
 | |
8BSM
 
 | | Human GLS in complex with compound 18 | | Descriptor: | 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial 65 kDa chain, ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.782 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSK
 
 | | Human GLS in complex with compound 3 | | Descriptor: | Glutaminase kidney isoform, mitochondrial 65 kDa chain, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide), ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSL
 
 | | Human GLS in complex with compound 12 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, ~{N}-[5-[[(3~{R})-1-(5-azanyl-1,3,4-thiadiazol-2-yl)pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]-2-phenyl-ethanamide | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
8BSN
 
 | | Human GLS in complex with compound 27 | | Descriptor: | (2~{S})-2-methoxy-2-phenyl-~{N}-[5-[[(3~{R})-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial 65 kDa chain, ... | | Authors: | Debreczeni, J.E. | | Deposit date: | 2022-11-25 | | Release date: | 2023-01-18 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Discovery of a Thiadiazole-Pyridazine-Based Allosteric Glutaminase 1 Inhibitor Series That Demonstrates Oral Bioavailability and Activity in Tumor Xenograft Models.
J Med Chem, 62, 2019
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
