6QH0
 
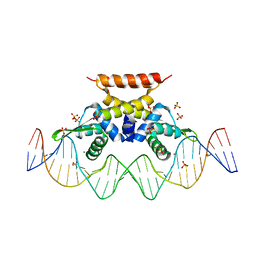 | | The complex structure of hsRosR-S5 (VNG0258H/RosR-S5) | | Descriptor: | DNA (28-MER), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QIL
 
 | | The complex structure of hsRosR-S1 (VNG0258H/RosR-S1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (28-MER), DNA binding protein, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-01-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
6QUA
 
 | | The complex structure of hsRosR-SG (vng0258/RosR-SG) | | Descriptor: | DNA (28-MER), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Shaanan, B, Kutnowski, N. | | Deposit date: | 2019-02-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Specificity of protein-DNA interactions in hypersaline environment: structural studies on complexes of Halobacterium salinarum oxidative stress-dependent protein hsRosR.
Nucleic Acids Res., 47, 2019
|
|
1T1M
 
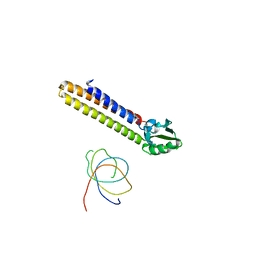 | | Binding position of ribosome recycling factor (RRF) on the E. coli 70S ribosome | | Descriptor: | 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA, ribosome recycling factor | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1T1O
 
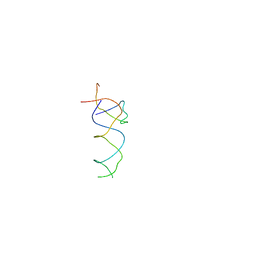 | | Components of the control 70S ribosome to provide reference for the RRF binding site | | Descriptor: | 19-mer fragment of the 23S rRNA, 42-mer fragment of double helix from 16S rRNA, dodecamer fragment of double helix from 23S rRNA | | Authors: | Agrawal, R.K, Sharma, M.R, Kiel, M.C, Hirokawa, G, Booth, T.M, Spahn, C.M, Grassucci, R.A, Kaji, A, Frank, J. | | Deposit date: | 2004-04-16 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6F5C
 
 | |
6EZ1
 
 | |
6FDH
 
 | |
6FAQ
 
 | |
5LXF
 
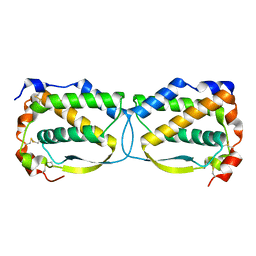 | | Crystal structure of the human Macrophage Colony Stimulating Factor M- CSF_C31S variant | | Descriptor: | Macrophage colony-stimulating factor 1 | | Authors: | Shahar, A, Papo, N, Zarivach, R, Kosloff, M, Bakhman, A, Rosenfeld, L, Zur, Y, Levaot, N. | | Deposit date: | 2016-09-21 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a monomeric variant of macrophage colony-stimulating factor (M-CSF) that antagonizes the c-FMS receptor.
Biochem. J., 474, 2017
|
|
