8UCI
 
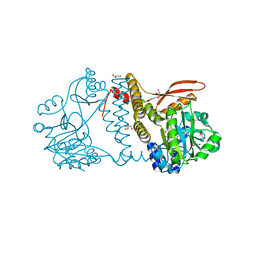 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
2ZEC
 
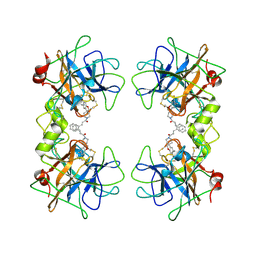 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-[1'-(3-phenylacryloyl)spiro[1-benzofuran-3,4'-piperidin]-5-yl]methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2ZEB
 
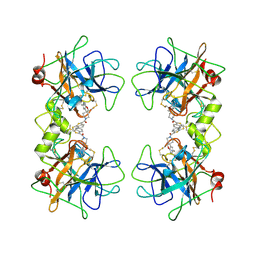 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | Descriptor: | 1-(1'-{[3-(methylsulfanyl)-2-benzothiophen-1-yl]carbonyl}spiro[1-benzofuran-3,4'-piperidin]-5-yl)methanamine, Tryptase beta 2 | | Authors: | Spurlino, J.C, Lewandowski, F, Milligan, C. | | Deposit date: | 2007-12-08 | | Release date: | 2008-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6MBI
 
 | |
3BD1
 
 | | Structure of the Cro protein from putative prophage element Xfaso 1 in Xylella fastidiosa strain Ann-1 | | Descriptor: | CHLORIDE ION, Cro protein, GLYCEROL, ... | | Authors: | Hall, B.M, Roberts, S.A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2007-11-13 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of
Cro proteins with 40% sequence identity but different folds
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6NPZ
 
 | | Crystal structure of Akt1 (aa 123-480) kinase with a bisubstrate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | Authors: | Chu, N, Cole, P.A, Gabelli, S.B. | | Deposit date: | 2019-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
6NXJ
 
 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6NSI
 
 | | Crystal structure of Fe(III)-bound YtgA from Chlamydia trachomatis | | Descriptor: | CALCIUM ION, FE (III) ION, Manganese-binding protein, ... | | Authors: | Luo, Z, Campbell, R, Begg, S.L, Kobe, B, McDevitt, C.A. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.00006342 Å) | | Cite: | Structure and Metal Binding Properties of Chlamydia trachomatis YtgA.
J.Bacteriol., 202, 2019
|
|
6NXI
 
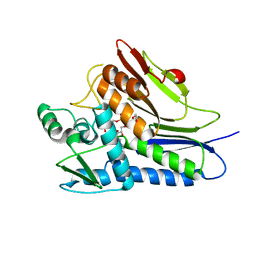 | | Flavin Transferase ApbE from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
3BO0
 
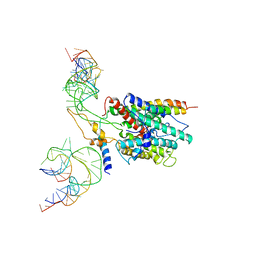 | | Ribosome-SecY complex | | Descriptor: | 23S RIBOSOMAL RNA, PREPROTEIN TRANSLOCASE SecE SUBUNIT, PREPROTEIN TRANSLOCASE SecY SUBUNIT, ... | | Authors: | Akey, C.W, Menetret, J.F. | | Deposit date: | 2007-12-15 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Ribosome binding of a single copy of the SecY complex: implications for protein translocation
Mol.Cell, 28, 2007
|
|
3BO1
 
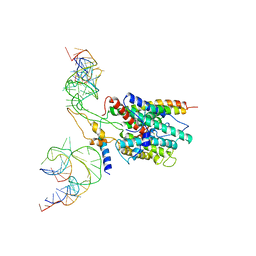 | | Ribosome-SecY complex | | Descriptor: | 23S RIBOSOMAL RNA, PREPROTEIN TRANSLOCASE SecE SUBUNIT, PREPROTEIN TRANSLOCASE SecY SUBUNIT, ... | | Authors: | Akey, C.W, Menetret, J.F. | | Deposit date: | 2007-12-15 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Ribosome binding of a single copy of the SecY complex: implications for protein translocation
Mol.Cell, 28, 2007
|
|
3D14
 
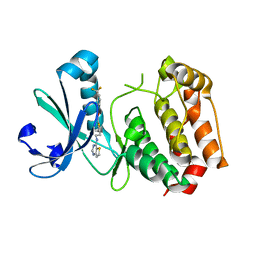 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)-ethyl]- thiazol-2-yl}-3-(3-trifluoromethyl-phenyl)-urea | | Descriptor: | 1-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}-3-[3-(trifluoromethyl)phenyl]urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Baskaran, S, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CW0
 
 | | E.coli DmsD | | Descriptor: | Twin-arginine leader-binding protein dmsD | | Authors: | Ramasamy, S, Clemons, W. | | Deposit date: | 2008-04-21 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the twin-arginine signal-binding protein DmsD from Escherichia coli.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3D15
 
 | | Crystal structure of mouse Aurora A (Asn186->Gly, Lys240->Arg, Met302->Leu) in complex with 1-(3-chloro-phenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)- ethyl]-thiazol-2-yl}-urea [SNS-314] | | Descriptor: | 1-(3-chlorophenyl)-3-{5-[2-(thieno[3,2-d]pyrimidin-4-ylamino)ethyl]-1,3-thiazol-2-yl}urea, serine/threonine kinase 6 | | Authors: | Elling, R.A, Bui, M, Allen, D.A, Oslob, J.D, Romanowski, M.J. | | Deposit date: | 2008-05-04 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a potent and selective aurora kinase inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3KA2
 
 | |
5FV0
 
 | | The cytoplasmic domain of EssC | | Descriptor: | 3'-O-[2-(METHYLAMINO)BENZOYL]ADENOSINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), ADENOSINE-5'-TRIPHOSPHATE, ESX secretion system protein EccC, ... | | Authors: | Zoltner, M, Ng, W.M.A.V, Palmer, T, Hunter, W.N. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | EssC: domain structures inform on the elusive translocation channel in the Type VII secretion system.
Biochem. J., 473, 2016
|
|
3J3I
 
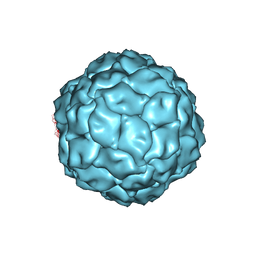 | | Penicillium chrysogenum virus (PcV) capsid structure | | Descriptor: | Capsid protein | | Authors: | Luque, D, Gomez-Blanco, J, Garriga, D, Brilot, A, Gonzalez, J.M, Havens, W.H, Carrascosa, J.L, Trus, B.L, Verdaguer, N, Grigorieff, N, Ghabrial, S.A, Caston, J.R. | | Deposit date: | 2013-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM near-atomic structure of a dsRNA fungal virus shows ancient structural motifs preserved in the dsRNA viral lineage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5G1Y
 
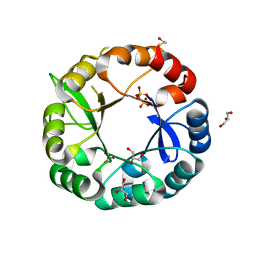 | | S. enterica HisA mutant D10G, dup13-15,V14:2M, Q24L, G102 | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-01 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G5J
 
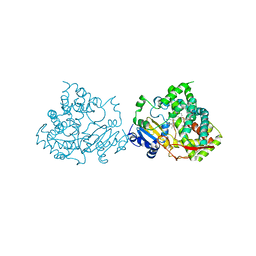 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
5GXG
 
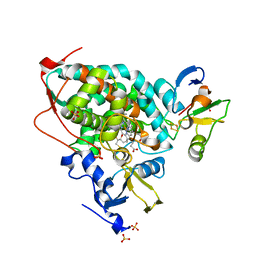 | | High-resolution crystal structure of the electron transfer complex of cytochrome p450cam with putidaredoxin | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kikui, Y, Hiruma, Y, Ubbink, M, Nojiri, M. | | Deposit date: | 2016-09-17 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of productive and futile encounters in an electron transfer protein complex
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G5I
 
 | | S. enterica HisA mutant D10G | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHOSPHATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-25 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2W
 
 | | S. enterica HisA mutant D10G, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-14 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G4E
 
 | | S. enterica HisA mutant D10G, Dup13-15, Q24L, G102A, V106L | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, ACETIC ACID, GLYCEROL, ... | | Authors: | Soderholm, A, Guo, X, Newton, M, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-05-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G1T
 
 | | S. enterica HisA mutant dup13-15, D10G | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHOSPHATE ION | | Authors: | Guo, X, Soderholm, A, Newton, M, Nasvall, J, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-03-30 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5G2H
 
 | | S. enterica HisA with mutation L169R | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, GLYCEROL, SODIUM ION, ... | | Authors: | Newton, M, Guo, X, Soderholm, A, Nasvall, J, Duarte, F, Andersson, D, Patrick, W, Selmer, M. | | Deposit date: | 2016-04-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
