1N4U
 
 | |
1N4W
 
 | |
4REK
 
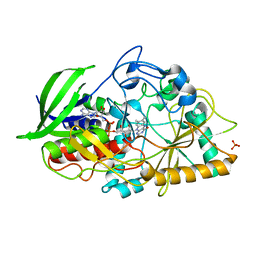 | | Crystal structure and charge density studies of cholesterol oxidase from Brevibacterium sterolicum at 0.74 ultra-high resolution | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zarychta, B, Lyubimov, A, Ahmed, M, Munshi, P, Guillot, B, Vrielink, A. | | Deposit date: | 2014-09-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Cholesterol oxidase: ultrahigh-resolution crystal structure and multipolar atom model-based analysis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8UD0
 
 | |
8UCZ
 
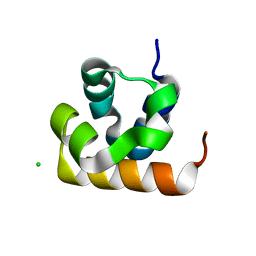 | |
8F2G
 
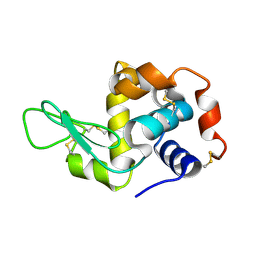 | | Crystal structure of Hen Egg White Lysozyme at 0.44 GPa | | Descriptor: | Lysozyme C | | Authors: | Marshall, A.C, Boer, S.A, Turner, G, Moggach, S.A, Bond, C.S, Vrielink, A. | | Deposit date: | 2022-11-08 | | Release date: | 2022-12-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | High-pressure single-crystal diffraction at the Australian Synchrotron.
J.Synchrotron Radiat., 30, 2023
|
|
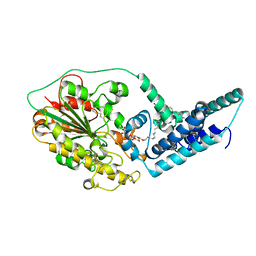
5FGN
 
 | |
5KWF
 
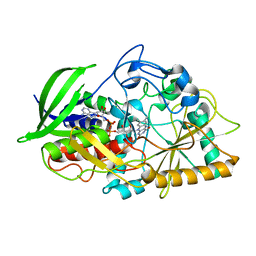 | | Joint X-ray Neutron Structure of Cholesterol Oxidase | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Golden, E, Vrielink, A, Meilleur, F, Blakeley, M. | | Deposit date: | 2016-07-18 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.499 Å), X-RAY DIFFRACTION | | Cite: | An extended N-H bond, driven by a conserved second-order interaction, orients the flavin N5 orbital in cholesterol oxidase.
Sci Rep, 7, 2017
|
|
1I19
 
 | | CRYSTAL STRUCTURE OF CHOLESTEROL OXIDASE FROM B.STEROLICUM | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHOLESTEROL OXIDASE, ... | | Authors: | Coulombe, R, Yue, K.Q, Ghisla, S, Vrielink, A. | | Deposit date: | 2001-01-31 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxygen access to the active site of cholesterol oxidase through a narrow channel is gated by an Arg-Glu pair.
J.Biol.Chem., 276, 2001
|
|
4XWR
 
 | |
4XXG
 
 | |
8UCY
 
 | |
4U2T
 
 | |
4U2L
 
 | | Dithionite reduced cholesterol in complex with sulfite | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, Cholesterol oxidase, SULFATE ION | | Authors: | Golden, E.A, Vrielink, A. | | Deposit date: | 2014-07-17 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.337 Å) | | Cite: | High-resolution structures of cholesterol oxidase in the reduced state provide insights into redox stabilization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4U2S
 
 | |
3GYJ
 
 | |
3GYI
 
 | |
1COY
 
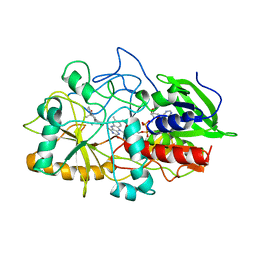 | | CRYSTAL STRUCTURE OF CHOLESTEROL OXIDASE COMPLEXED WITH A STEROID SUBSTRATE. IMPLICATIONS FOR FAD DEPENDENT ALCOHOL OXIDASES | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, CHOLESTEROL OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, J, Vrielink, A, Brick, P, Blow, D.M. | | Deposit date: | 1993-06-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cholesterol oxidase complexed with a steroid substrate: implications for flavin adenine dinucleotide dependent alcohol oxidases.
Biochemistry, 32, 1993
|
|
3CNJ
 
 | |
1QD1
 
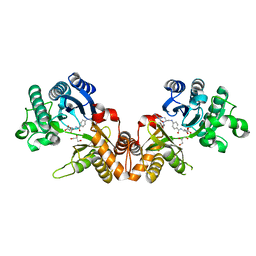 | | THE CRYSTAL STRUCTURE OF THE FORMIMINOTRANSFERASE DOMAIN OF FORMIMINOTRANSFERASE-CYCLODEAMINASE. | | Descriptor: | FORMIMINOTRANSFERASE-CYCLODEAMINASE, GLYCEROL, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid | | Authors: | Kohls, D, Sulea, T, Purisima, E, MacKenzie, R.E, Vrielink, A. | | Deposit date: | 1999-07-08 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the formiminotransferase domain of formiminotransferase-cyclodeaminase: implications for substrate channeling in a bifunctional enzyme.
Structure Fold.Des., 8, 2000
|
|
3B6D
 
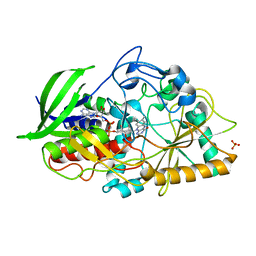 | |
3B3R
 
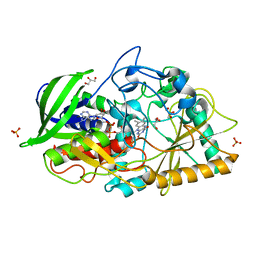 | | Crystal structure of Streptomyces cholesterol oxidase H447Q/E361Q mutant bound to glycerol (0.98A) | | Descriptor: | Cholesterol oxidase, FLAVIN-N7 PROTONATED-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lyubimov, A.Y, Heard, K, Tang, H, Sampson, N.S, Vrielink, A. | | Deposit date: | 2007-10-22 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Distortion of flavin geometry is linked to ligand binding in cholesterol oxidase
Protein Sci., 16, 2007
|
|
2IID
 
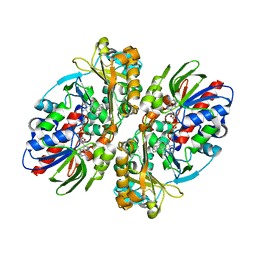 | | Structure of L-amino acid oxidase from Calloselasma rhodostoma in complex with L-phenylalanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino-acid oxidase, ... | | Authors: | Moustafa, I.M, Foster, S, Lyubimov, A.Y, Vrielink, A. | | Deposit date: | 2006-09-27 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of LAAO from Calloselasma rhodostoma with an L-Phenylalanine Substrate: Insights into Structure and Mechanism
J.Mol.Biol., 364, 2006
|
|
1EZ0
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | Descriptor: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
1EYY
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | Descriptor: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | Deposit date: | 2000-05-09 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
