4DN4
 
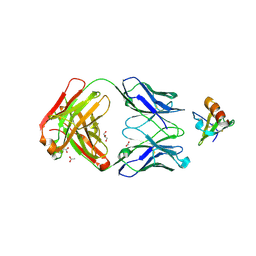 | | Crystal structure of the complex between cnto888 fab and mcp-1 mutant p8a | | Descriptor: | ACETATE ION, C-C motif chemokine 2, CNTO888 HEAVY CHAIN, ... | | Authors: | Obmolova, G, Teplyakov, A, Malia, T, Grygiel, T, Sweet, R, Snyder, L, Gilliland, G. | | Deposit date: | 2012-02-08 | | Release date: | 2012-10-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for high selectivity of anti-CCL2 neutralizing antibody CNTO 888.
Mol.Immunol., 51, 2012
|
|
1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
3QPX
 
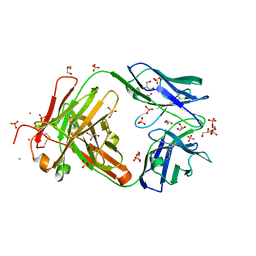 | | Crystal structure of Fab C2507 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab C2507 heavy chain, ... | | Authors: | Luo, J, Teplyakov, A, Obmolova, O, Malia, T, Gilliland, G.L. | | Deposit date: | 2011-02-14 | | Release date: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Fab C2507
To be Published
|
|
3QPQ
 
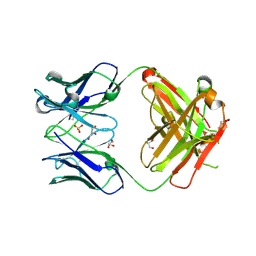 | | Crystal structure of ANTI-TLR3 antibody C1068 FAB | | Descriptor: | C1068 heavy chain, C1068 light chain, GLYCEROL, ... | | Authors: | Luo, J, Obmolova, G, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2011-02-14 | | Release date: | 2011-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Fab C1068
To be Published
|
|
3TES
 
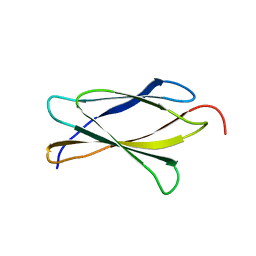 | | Crystal Structure of Tencon | | Descriptor: | Tencon | | Authors: | Luo, J, Jacobs, S, Teplyakov, A, Obmolova, G, O'Neil, K, Gilliland, G. | | Deposit date: | 2011-08-15 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of novel FN3 domains with high stability by a consensus sequence approach.
Protein Eng.Des.Sel., 25, 2012
|
|
3TEU
 
 | | Crystal structure of fibcon | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Fibcon | | Authors: | Luo, J, Jacobs, S, Teplyakov, A, Obmolova, G, O'Neil, K, Gilliland, G. | | Deposit date: | 2011-08-15 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.002 Å) | | Cite: | Design of novel FN3 domains with high stability by a consensus sequence approach.
Protein Eng.Des.Sel., 25, 2012
|
|
3ULS
 
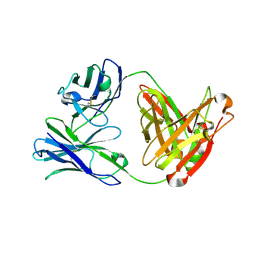 | | Crystal structure of Fab12 | | Descriptor: | Fab12 heavy chain, Fab12 light chain | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3ULU
 
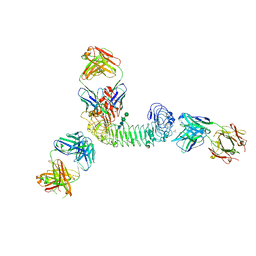 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3ULV
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.522 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
1E9G
 
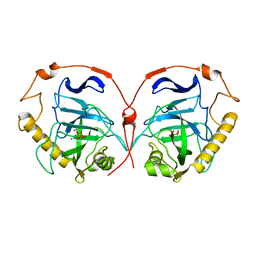 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Heikinheimo, P, Tuominen, V, Ahonen, A.-K, Teplyakov, A, Cooperman, B.S, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2000-10-12 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Toward a Quantum-Mechanical Description of Metal-Assisted Phosphoryl Transfer in Pyrophosphatase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1E6A
 
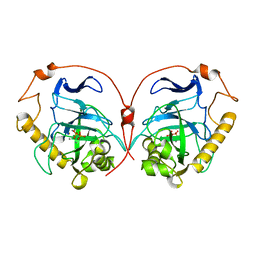 | | Fluoride-inhibited substrate complex of Saccharomyces cerevisiae inorganic pyrophosphatase | | Descriptor: | FLUORIDE ION, INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Heikinheimo, P, Tuominen, V, Ahonen, A.-K, Teplyakov, A, Cooperman, B.S, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2000-08-09 | | Release date: | 2001-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward a quantum-mechanical description of metal-assisted phosphoryl transfer in pyrophosphatase.
Proc. Natl. Acad. Sci. U.S.A., 98, 2001
|
|
1LQA
 
 | | TAS PROTEIN FROM ESCHERICHIA COLI IN COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tas protein | | Authors: | Obmolova, G, Teplyakov, A, Khil, P.P, Howard, A.J, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1NIJ
 
 | | YJIA PROTEIN | | Descriptor: | Hypothetical protein yjiA | | Authors: | Khil, P.P, Obmolova, G, Teplyakov, A, Howard, A.J, Gilliland, G.L, Camerini-Otero, R.D, Structure 2 Function Project (S2F) | | Deposit date: | 2002-12-24 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Escherichia coli YjiA protein suggests a GTP-dependent regulatory function.
Proteins, 54, 2004
|
|
1NMO
 
 | | Structural genomics, protein ybgI, unknown function | | Descriptor: | FE (III) ION, Hypothetical protein ybgI | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1NMP
 
 | | Structural genomics, ybgI protein, unknown function | | Descriptor: | Hypothetical protein ybgI, MAGNESIUM ION | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
4M6A
 
 | | N-Terminal beta-Strand Swapping in a Consensus Derived Alternative Scaffold Driven by Stabilizing Hydrophobic Interactions | | Descriptor: | Tencon | | Authors: | Luo, J, Teplyakov, A, Obmolova, G, Malia, T.J, Chan, W, Jocobs, S.A, O'neil, K.T, Gilliland, G.L. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | N-terminal beta-strand swapping in a consensus-derived alternative scaffold driven by stabilizing hydrophobic interactions.
Proteins, 82, 2014
|
|
4MA3
 
 | | Crystal structure of anti-hinge rabbit antibody C2095 | | Descriptor: | ACETATE ION, C2095 heavy chain, C2095 light chain, ... | | Authors: | Malia, T, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2013-08-15 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
1JJV
 
 | | DEPHOSPHO-COA KINASE IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEPHOSPHO-COA KINASE, MERCURY (II) ION, ... | | Authors: | Obmolova, G, Teplyakov, A, Bonander, N, Eisenstein, E, Howard, A.J, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-09 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dephospho-coenzyme A kinase from Haemophilus influenzae.
J.Struct.Biol., 136, 2001
|
|
4O51
 
 | | Crystal structure of the QAA variant of anti-hinge rabbit antibody 2095-2 in complex with IDES hinge peptide | | Descriptor: | IDES hinge peptide, QAA-2095-2 heavy chain, QAA-2095-2 light chain | | Authors: | Malia, T.J, Luo, J, Teplyakov, A, Gilliland, G.L. | | Deposit date: | 2013-12-19 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structure and specificity of an antibody targeting a proteolytically cleaved IgG hinge.
Proteins, 82, 2014
|
|
1DPC
 
 | |
1DPB
 
 | |
1DPD
 
 | |
1YPP
 
 | | ACID ANHYDRIDE HYDROLASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Harutyunyan, E.H, Kuranova, I.P, Lamzin, V.S, Dauter, Z, Wilson, K.S. | | Deposit date: | 1996-05-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of yeast inorganic pyrophosphatase complexed with manganese and phosphate.
Eur.J.Biochem., 239, 1996
|
|
8DY2
 
 | | Crystal Structure of spFv GLK1 | | Descriptor: | SULFATE ION, spFv GLK1 LH | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
8DY4
 
 | | Crystal Structure of spFv CAT2200 HL | | Descriptor: | spFv CAT2200 HL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2022-08-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | "Stapling" scFv for multispecific biotherapeutics of superior properties.
Mabs, 15, 2023
|
|
