5KVH
 
 | | Crystal structure of human apoptosis-inducing factor with W196A mutation | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
5KVI
 
 | | Crystal structure of monomeric human apoptosis-inducing factor with E413A/R422A/R430A mutations | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Apoptosis-inducing factor 1, mitochondrial, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
7N8W
 
 | |
7N8V
 
 | |
2SSP
 
 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
1UGH
 
 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | Descriptor: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
5UM9
 
 | | Flap endonuclease 1 (FEN1) D86N with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
8D3G
 
 | |
8D3J
 
 | |
8D3O
 
 | |
8D3E
 
 | |
8D3H
 
 | |
8D3I
 
 | |
8D3N
 
 | |
8D3K
 
 | |
3UMV
 
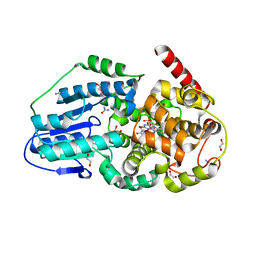 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2011-11-14 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
3UEK
 
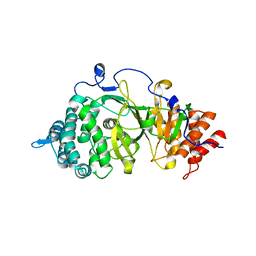 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase | | Descriptor: | Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEL
 
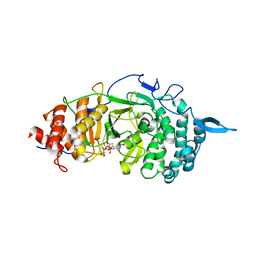 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase bound to ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Poly(ADP-ribose) glycohydrolase | | Authors: | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | Deposit date: | 2011-10-30 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VHJ
 
 | |
1QOM
 
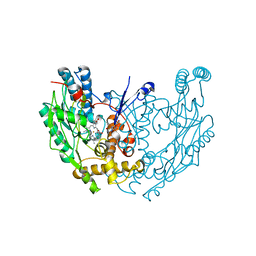 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH SWAPPED N-TERMINAL HOOK | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Crane, B.R, Rosenfeld, R.A, Arvai, A.S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-15 | | Release date: | 1999-12-15 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-Terminal Domain Swapping and Metal Ion Binding in Nitric Oxide Synthase Dimerization
Embo J., 18, 1999
|
|
1DWX
 
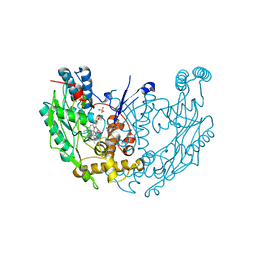 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DWV
 
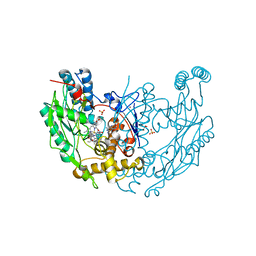 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DWW
 
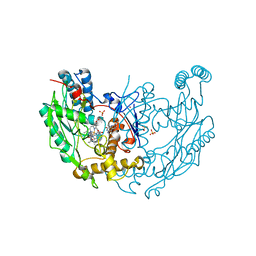 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and dihydrobiopterin | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
2AWM
 
 | | GFP R96A chromophore maturation recovery mutant R96A Q183R | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
2AWK
 
 | | GFP R96M mature chromophore | | Descriptor: | MAGNESIUM ION, green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis.
Biochemistry, 44, 2005
|
|
