3JT1
 
 | | Legionella pneumophila glucosyltransferase Lgt1, UDP-bound form | | Descriptor: | Putative uncharacterized protein, URIDINE-5'-DIPHOSPHATE | | Authors: | Lu, W, Du, J, Belyi, Y, Stahl, M, Zivilikidis, T, Gerhardt, S, Aktories, K, Einsle, O. | | Deposit date: | 2009-09-11 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
J.Mol.Biol., 2009
|
|
3JSZ
 
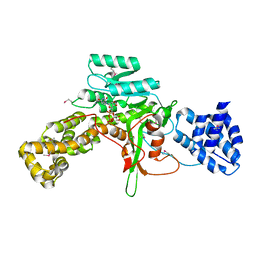 | | Legionella pneumophila glucosyltransferase Lgt1 N293A with UDP-Glc | | Descriptor: | MAGNESIUM ION, Putative uncharacterized protein, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lu, W, Du, J, Belyi, Y, Stahl, M, Zivilikidis, T, Gerhardt, S, Aktories, K, Einsle, O. | | Deposit date: | 2009-09-11 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of the Action of Glucosyltransferase Lgt1 from Legionella pneumophila.
J.Mol.Biol., 2009
|
|
5UP2
 
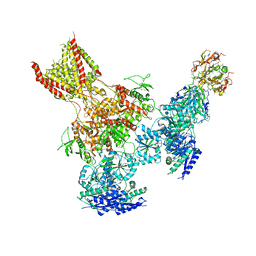 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, Ro 25-6981, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GluN2B-specific Fab, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
5UOW
 
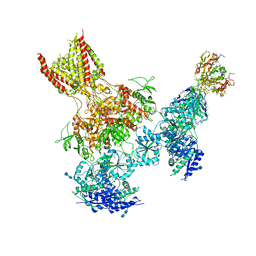 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2B in complex with glycine, glutamate, MK-801 and a GluN2B-specific Fab, at pH 6.5 | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Lu, W, Du, J, Goehring, A, Gouaux, E. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structures of the triheteromeric NMDA receptor and its allosteric modulation.
Science, 355, 2017
|
|
6UIW
 
 | |
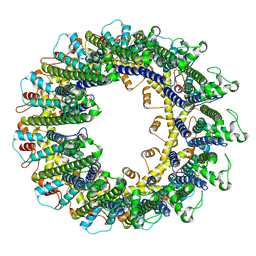
6UIV
 
 | |
6UIX
 
 | | Cryo-EM structure of human CALHM2 gap junction | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Lu, W, Du, J, Choi, W. | | Deposit date: | 2019-10-01 | | Release date: | 2019-11-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structures and gating mechanism of human calcium homeostasis modulator 2.
Nature, 576, 2019
|
|
6WBK
 
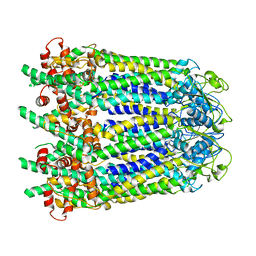 | |
6WBN
 
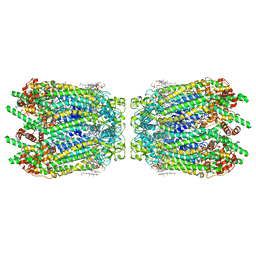 | | Cryo-EM structure of human Pannexin 1 channel N255A mutant, gap junction | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, DIACYL GLYCEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBM
 
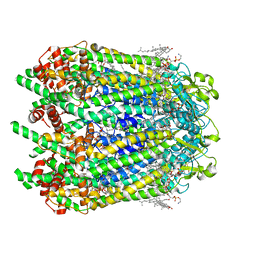 | | Cryo-EM structure of human Pannexin 1 channel N255A mutant | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL, DIACYL GLYCEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBL
 
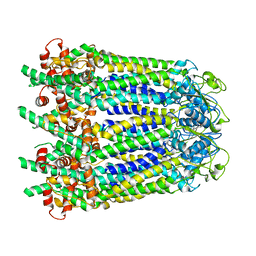 | |
6WBG
 
 | | Cryo-EM structure of human Pannexin 1 channel with its C-terminal tail cleaved by caspase-7 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBI
 
 | |
6WBF
 
 | | Cryo-EM structure of wild type human Pannexin 1 channel | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6CUD
 
 | | Structure of the human TRPC3 in a lipid-occupied, closed state | | Descriptor: | (2R)-3-hydroxypropane-1,2-diyl dihexanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, W, Du, J, Fan, C, Choi, W. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human lipid-gated cation channel TRPC3.
Elife, 7, 2018
|
|
7JNA
 
 | | Cryo-EM structure of human proton-activated chloride channel PAC at pH 8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Proton-activated chloride channel | | Authors: | Lu, W, Ruan, R, Du, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and pH-sensing mechanism of the proton-activated chloride channel.
Nature, 588, 2020
|
|
7JNC
 
 | | cryo-EM structure of human proton-activated chloride channel PAC at pH 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Proton-activated chloride channel | | Authors: | Lu, W, Ruan, R, Du, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structures and pH-sensing mechanism of the proton-activated chloride channel.
Nature, 588, 2020
|
|
2ZJL
 
 | | Crystal structure of the human BACE1 catalytic domain in complex with N-[1-(5-bromo-2,3-dimethoxy-benzyl)-piperidin-4-yl]-4-mercapto-butyramide | | Descriptor: | Beta-secretase 1, N-[1-(5-bromo-2,3-dimethoxybenzyl)piperidin-4-yl]-4-sulfanylbutanamide | | Authors: | Randal, M, Lam, M.B, Lu, W, Romanowski, M.J. | | Deposit date: | 2008-03-07 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based discovery of novel BACE1 inhibitors using Tethering technology
To be Published
|
|
2ZJK
 
 | |
1CT4
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-VAL18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CSO
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-ILE18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT0
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-SER18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CT2
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-THR18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
1F3M
 
 | | CRYSTAL STRUCTURE OF HUMAN SERINE/THREONINE KINASE PAK1 | | Descriptor: | IODIDE ION, SERINE/THREONINE-PROTEIN KINASE PAK-ALPHA | | Authors: | Lei, M, Lu, W, Meng, W, Parrini, M.-C, Eck, M.J, Mayer, B.J, Harrison, S.C. | | Deposit date: | 2000-06-05 | | Release date: | 2000-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch.
Cell(Cambridge,Mass.), 102, 2000
|
|
