6MTS
 
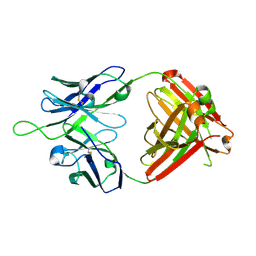 | | Crystal structure of VRC43.03 Fab | | Descriptor: | Antibody VRC43.03 Fab heavy chain, Antibody VRC43.03 Fab light chain | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.437 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
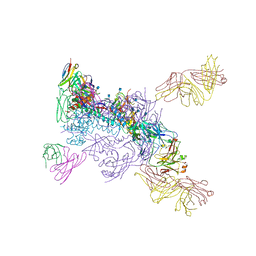
6MU7
 
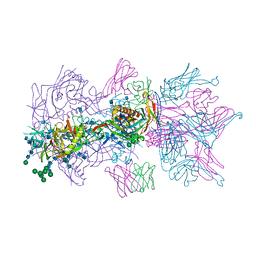 | |
6MUG
 
 | |
6MJZ
 
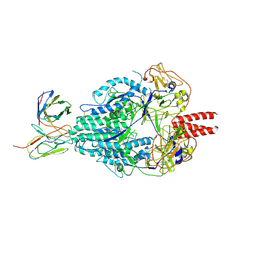 | | Cryo-EM structure of Human Parainfluenza Virus Type 3 (hPIV3) in complex with antibody PIA174 | | Descriptor: | Fusion glycoprotein F0, PIA174 Fab Heavy chain, PIA174 Fab Light chain | | Authors: | Acharya, P, Stewart-Jones, G, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6MQM
 
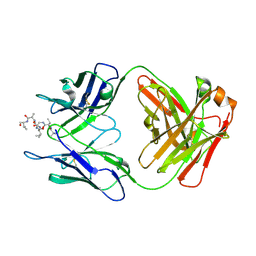 | |
6MYY
 
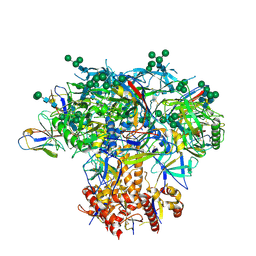 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 3 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-02 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6MTJ
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-378806 in Complex with Human Antibodies 3H109L and 35O22 at 2.9 Angstrom | | Descriptor: | 1-[(2R)-4-(benzenecarbonyl)-2-methylpiperazin-1-yl]-2-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)ethane-1,2-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-19 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6MTP
 
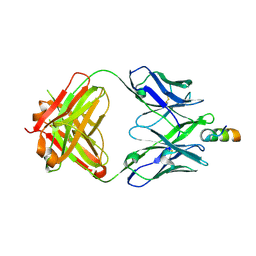 | | Crystal structure of VRC42.04 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC42.04 Fab heavy chain, Antibody VRC42.04 Fab light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MQR
 
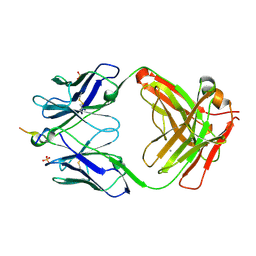 | | Vaccine-elicited NHP FP-targeting neutralizing antibody 0PV-a.01 in complex with FP (residue 512-519) | | Descriptor: | CALCIUM ION, HIV fusion peptide residue 512-519, SULFATE ION, ... | | Authors: | Xu, K, Wang, Y, Kwong, P.D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
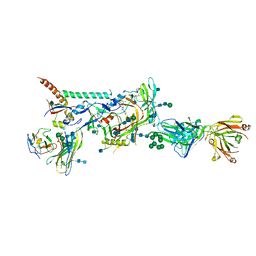
6MUF
 
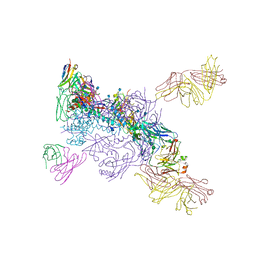 | |
6MTN
 
 | |
6MTT
 
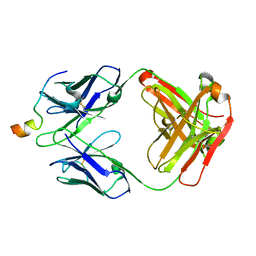 | | Crystal structure of VRC46.01 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC46.01 Fab heavy chain, Antibody VRC46.01 Fb light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MU8
 
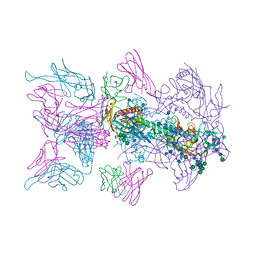 | |
6N16
 
 | |
6N1W
 
 | |
6N1V
 
 | |
6MTQ
 
 | | Crystal structure of VRC42.N1 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.N1 Fab heavy chain, Antibody VRC42.N1 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Law, W.H, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6MU6
 
 | | Crystal Structure of HIV-1 BG505 SOSIP.664 Prefusion Env Trimer Bound to Small Molecule HIV-1 Entry Inhibitor BMS-814508 in Complex with Human Antibodies 3H109L and 35O22 at 3.2 Angstrom | | Descriptor: | (2R)-{1-[{7-[2-({[3-(dimethylamino)propyl](methyl)amino}methyl)-1,3-thiazol-4-yl]-4-methoxy-1H-pyrrolo[2,3-c]pyridin-3-yl}(oxo)acetyl]piperidin-4-yl}(phenyl)acetonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, Y.-T, Kwong, P.D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Lattice engineering enables definition of molecular features allowing for potent small-molecule inhibition of HIV-1 entry.
Nat Commun, 10, 2019
|
|
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MQE
 
 | |
6MQS
 
 | |
6MZJ
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
6NNJ
 
 | |
6NNF
 
 | |
6NF2
 
 | |
