1Z2F
 
 | | solution structure of CfAFP-501 | | Descriptor: | Antifreeze Protein Isoform 501 | | Authors: | Li, C, Jin, C. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of an Antifreeze Protein CfAFP-501 from Choristoneura fumiferana
J.Biomol.Nmr, 32, 2005
|
|
2B59
 
 | |
6KJB
 
 | | wild-type apo-form E. coli ATCase holoenzyme with an unusual open conformation of R167 | | Descriptor: | Aspartate carbamoyltransferase catalytic subunit, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Wang, N, Lei, Z, Zheng, J, Jia, Z.C. | | Deposit date: | 2019-07-22 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Conformational Plasticity of the Active Site Entrance inE. coliAspartate Transcarbamoylase and Its Implication in Feedback Regulation.
Int J Mol Sci, 21, 2020
|
|
4GKP
 
 | |
4GKQ
 
 | |
4GKR
 
 | |
7Q3V
 
 | |
1EEN
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-D-A-D-BPA-PTYR-L-I-P-Q-Q-G | | Descriptor: | ACETIC ACID, ALA-ASP-PBF-PTR-LEU-ILE-PRO, MAGNESIUM ION, ... | | Authors: | Puius, Y.A, Zhao, Y, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
1EEO
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 | | Descriptor: | ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 PEPTIDE, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Sarmiento, M, Puius, Y.A, Vetter, S.W, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
6LJ9
 
 | |
6LJB
 
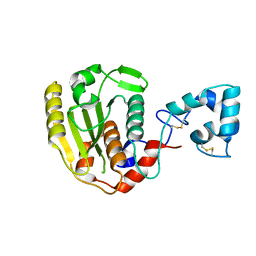 | |
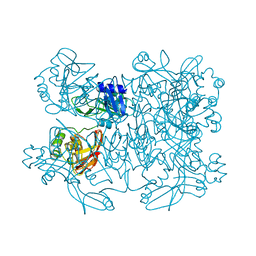
5YVD
 
 | |
1QFR
 
 | | NMR SOLUTION STRUCTURE OF PHOSPHOCARRIER PROTEIN HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Maurer, T, Doeker, R, Goerler, A, Hengstenberg, W, Kalbitzer, H.R. | | Deposit date: | 1999-04-13 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the histidine-containing phosphocarrier protein (HPr) from Enterococcus faecalis in solution.
Eur.J.Biochem., 268, 2001
|
|
4P69
 
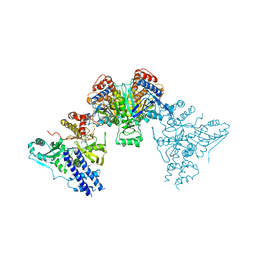 | | Acek (D477A) ICDH complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NADP], ... | | Authors: | Jimin, Z, Nan, W, Shu, W, Zongchao, J. | | Deposit date: | 2014-03-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The phosphatase mechanism of bifunctional kinase/phosphatase AceK.
Chem. Commun. (Camb.), 50, 2014
|
|
1TL9
 
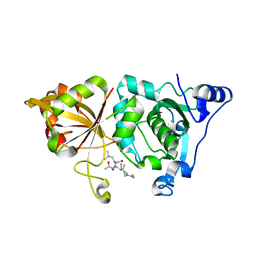 | | High resolution crystal structure of calpain I protease core in complex with leupeptin | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
1TLO
 
 | | High resolution crystal structure of calpain I protease core in complex with E64 | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Moldoveanu, T, Campbell, R.L, Cuerrier, D, Davies, P.L. | | Deposit date: | 2004-06-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Calpain-E64 and -Leupeptin Inhibitor Complexes Reveal Mobile Loops Gating the Active Site
J.Mol.Biol., 343, 2004
|
|
3DMS
 
 | |
6KZ6
 
 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
5XDM
 
 | |
7E3O
 
 | |
5Y4Z
 
 | | Crystal structure of the Zika virus NS3 helicase complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, L. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural view of the helicase reveals that Zika virus uses a conserved mechanism for unwinding RNA
Acta Crystallogr.,Sect.F, 74, 2018
|
|
7D9M
 
 | | grass carp interleukin-2 | | Descriptor: | Interleukin | | Authors: | Junya, w, Jun, z. | | Deposit date: | 2020-10-13 | | Release date: | 2020-10-28 | | Last modified: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the co-evolution of IL-2 and its private receptor in fish.
Dev.Comp.Immunol., 115, 2020
|
|
1AAX
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO BIS(PARA-PHOSPHOPHENYL)METHANE (BPPM) MOLECULES | | Descriptor: | 4-PHOSPHONOOXY-PHENYL-METHYL-[4-PHOSPHONOOXY]BENZEN, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Puius, Y.A, Zhao, Y, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1C8A
 
 | |
1C89
 
 | |
