1B7I
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 K61R | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1B7K
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 R47H | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1B7J
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 V20A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1EKL
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 E35K | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-21 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
9AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 S42G | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
9MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18N | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1MSJ
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
5MSI
 
 | |
1JAB
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
7MSI
 
 | |
2D40
 
 | |
2F2O
 
 | |
2F2P
 
 | |
2FSU
 
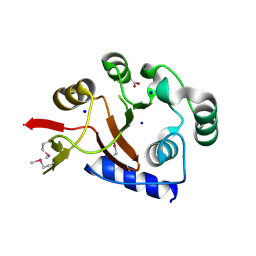 | | Crystal Structure of the PhnH Protein from Escherichia Coli | | Descriptor: | ACETATE ION, Protein phnH, SODIUM ION | | Authors: | Adams, M.A, Luo, Y, Zechel, D.L, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-23 | | Release date: | 2007-03-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of PhnH: an essential component of carbon-phosphorus lyase in Escherichia coli.
J.Bacteriol., 190, 2008
|
|
2R19
 
 | |
2R1A
 
 | |
2R28
 
 | |
1MDW
 
 | | Crystal Structure of Calcium-Bound Protease Core of Calpain II Reveals the Basis for Intrinsic Inactivation | | Descriptor: | CALCIUM ION, Calpain II, catalytic subunit | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Jia, Z, Davies, P.L. | | Deposit date: | 2002-08-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Calpain silencing by a reversible intrinsic mechanism.
Nat.Struct.Biol., 10, 2003
|
|
4MSI
 
 | |
1XZO
 
 | | Identification of a disulfide switch in BsSco, a member of the Sco family of cytochrome c oxidase assembly proteins | | Descriptor: | CADMIUM ION, CALCIUM ION, Hypothetical protein ypmQ | | Authors: | Ye, Q, Imriskova-Sosova, I, Hill, B.C, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Identification of a Disulfide Switch in BsSco, a Member of the Sco Family of Cytochrome c Oxidase Assembly Proteins
Biochemistry, 44, 2005
|
|
1ZYL
 
 | |
2MSI
 
 | |
2B3D
 
 | |
2AMJ
 
 | |
1DKM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
