1PBD
 
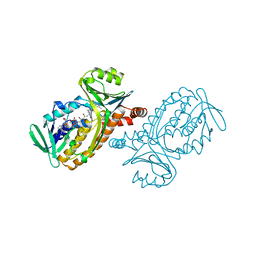 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 4-AMINOBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1PBB
 
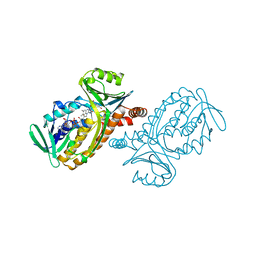 | | CRYSTAL STRUCTURES OF WILD-TYPE P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-AMINOBENZOATE, 2,4-DIHYDROXYBENZOATE AND 2-HYDROXY-4-AMINOBENZOATE AND OF THE TRY222ALA MUTANT, COMPLEXED WITH 2-HYDROXY-4-AMINOBENZOATE. EVIDENCE FOR A PROTON CHANNEL AND A NEW BINDING MODE OF THE FLAVIN RING | | Descriptor: | 2,4-DIHYDROXYBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE | | Authors: | Schreuder, H.A, Mattevi, A, Hol, W.G.J. | | Deposit date: | 1994-07-06 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of wild-type p-hydroxybenzoate hydroxylase complexed with 4-aminobenzoate,2,4-dihydroxybenzoate, and 2-hydroxy-4-aminobenzoate and of the Tyr222Ala mutant complexed with 2-hydroxy-4-aminobenzoate. Evidence for a proton channel and a new binding mode of the flavin ring
Biochemistry, 33, 1994
|
|
1QB5
 
 | |
1HCY
 
 | |
1HC1
 
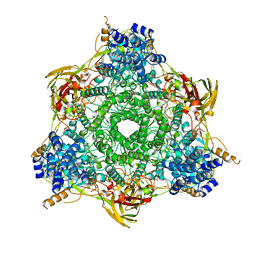 | |
1PBE
 
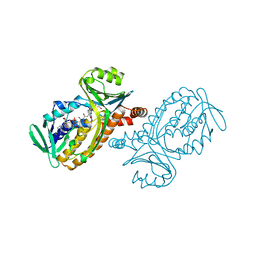 | |
1BTM
 
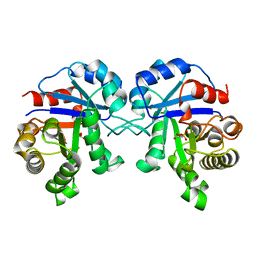 | | TRIOSEPHOSPHATE ISOMERASE (TIM) COMPLEXED WITH 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Delboni, L.F, Mande, S.C, Hol, W.G.J. | | Deposit date: | 1995-11-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions.
Protein Sci., 4, 1995
|
|
1ATH
 
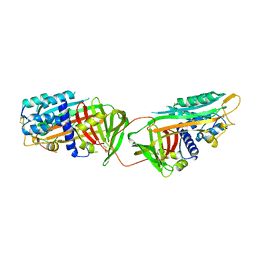 | |
1RL4
 
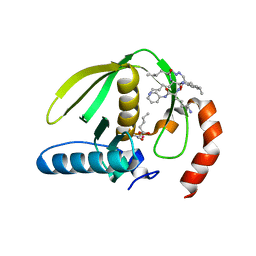 | | Plasmodium falciparum peptide deformylase complex with inhibitor | | Descriptor: | (2R)-2-{[FORMYL(HYDROXY)AMINO]METHYL}HEXANOIC ACID, 2-{N'-[2-(5-AMINO-1-PHENYLCARBAMOYL-PENTYLCARBAMOYL)-HEXYL]-HYDRAZINOMETHYL}-HEXANOIC ACID(5-AMINO-1-PHENYLCARBAMOYL-PENTYL)-AMIDE, COBALT (II) ION, ... | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G.J. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase.
Protein Sci., 13, 2004
|
|
5TIM
 
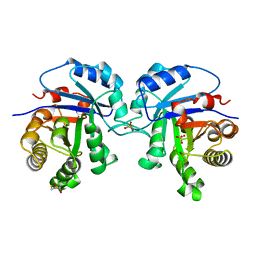 | | REFINED 1.83 ANGSTROMS STRUCTURE OF TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE, CRYSTALLIZED IN THE PRESENCE OF 2.4 M-AMMONIUM SULPHATE. A COMPARISON WITH THE STRUCTURE OF THE TRYPANOSOMAL TRIOSEPHOSPHATE ISOMERASE-GLYCEROL-3-PHOSPHATE COMPLEX | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Wierenga, R.K, Hol, W.G.J. | | Deposit date: | 1991-04-23 | | Release date: | 1992-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex.
J.Mol.Biol., 220, 1991
|
|
3BWC
 
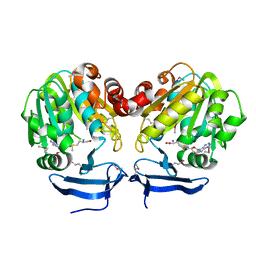 | | Crystal structure of spermidine synthase from Trypanosoma cruzi in complex with SAM at 2.3 A resolution | | Descriptor: | BETA-MERCAPTOETHANOL, S-ADENOSYLMETHIONINE, Spermidine synthase | | Authors: | Bosch, J, Arakaki, T.L, Le Trong, I, Merritt, E.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2008-01-09 | | Release date: | 2008-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of spermidine synthase from Trypanosoma cruzi.
To be Published
|
|
1CHQ
 
 | |
1CHP
 
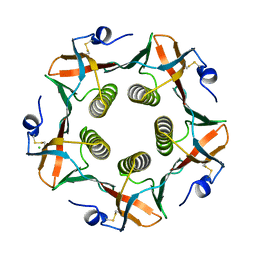 | |
1EEI
 
 | |
1U8R
 
 | | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions | | Descriptor: | COBALT (II) ION, Iron-dependent repressor ideR, SODIUM ION, ... | | Authors: | Wisedchaisri, G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of an IdeR-DNA Complex Reveals a Conformational Change in Activated IdeR for Base-specific Interactions.
J.Mol.Biol., 342, 2004
|
|
1HTL
 
 | |
1SYR
 
 | |
1TC5
 
 | |
1UV7
 
 | | periplasmic domain of EpsM from Vibrio cholerae | | Descriptor: | GENERAL SECRETION PATHWAY PROTEIN M | | Authors: | Abendroth, J, Hol, W.G.J. | | Deposit date: | 2004-01-15 | | Release date: | 2004-04-23 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Periplasmic Domain of the Type II Secretion System Protein Epsm from Vibrio Cholerae: The Simplest Version of the Ferredoxin Fold
J.Mol.Biol., 338, 2004
|
|
1MAE
 
 | |
1MAF
 
 | |
1NOP
 
 | | Crystal structure of human tyrosyl-DNA phosphodiesterase (Tdp1) in complex with vanadate, DNA and a human topoisomerase I-derived peptide | | Descriptor: | 5'-D(*AP*GP*AP*GP*TP*T)-3', VANADATE ION, topoisomerase I-derived peptide, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 2003-01-16 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a transition state mimic for Tdp1 assembled from vanadate, DNA, and a topoisomerase I-derived peptide
Chem.Biol., 10, 2003
|
|
1DPB
 
 | |
1DPD
 
 | |
1DPC
 
 | |
