3N4D
 
 | |
2GDG
 
 | |
4LHP
 
 | | Crystal Structure of Native FG41Malonate Semialdehyde Decarboxylase | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
4LHO
 
 | | Crystal Structure of FG41Malonate Semialdehyde Decarboxylase inhibited by 3-bromopropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
2AAJ
 
 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
2AAL
 
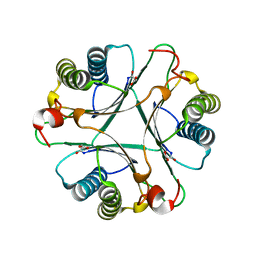 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | MALONATE ION, Malonate semialdehyde decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
2AAG
 
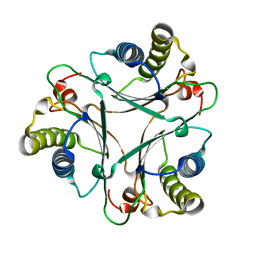 | | Crystal Structures of the Wild-type, Mutant-P1A and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities | | Descriptor: | Malonate Semialdehyde Decarboxylase | | Authors: | Almrud, J.J, Poelarends, G.J, Johnson Jr, W.H, Serrano, H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2005-07-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of the Wild-Type, P1A Mutant, and Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis for the Decarboxylase and Hydratase Activities
Biochemistry, 44, 2005
|
|
2FM7
 
 | |
1E2O
 
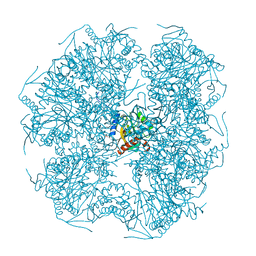 | | CATALYTIC DOMAIN FROM DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE | | Descriptor: | DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE, SULFATE ION | | Authors: | Knapp, J.E, Mitchell, D.T, Yazdi, M.A, Ernst, S.R, Reed, L.J, Hackert, M.L. | | Deposit date: | 1998-05-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex.
J.Mol.Biol., 280, 1998
|
|
1BJP
 
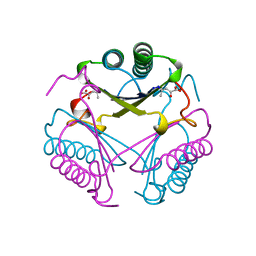 | | CRYSTAL STRUCTURE OF 4-OXALOCROTONATE TAUTOMERASE INACTIVATED BY 2-OXO-3-PENTYNOATE AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | 2-OXO-3-PENTENOIC ACID, 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Taylor, A.B, Czerwinski, R.M, Johnson Junior, W.H, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-06-26 | | Release date: | 1998-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 4-oxalocrotonate tautomerase inactivated by 2-oxo-3-pentynoate at 2.4 A resolution: analysis and implications for the mechanism of inactivation and catalysis.
Biochemistry, 37, 1998
|
|
1D7K
 
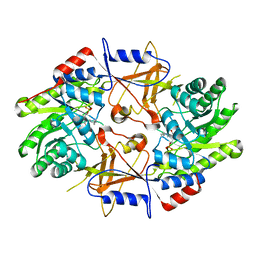 | | CRYSTAL STRUCTURE OF HUMAN ORNITHINE DECARBOXYLASE AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN ORNITHINE DECARBOXYLASE | | Authors: | Almrud, J.J, Oliveira, M.A, Kern, A.D, Grishin, N.V, Phillips, M.A, Hackert, M.L. | | Deposit date: | 1999-10-18 | | Release date: | 2000-10-25 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human ornithine decarboxylase at 2.1 A resolution: structural insights to antizyme binding.
J.Mol.Biol., 295, 2000
|
|
1HBR
 
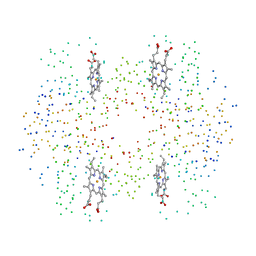 | | R-STATE FORM OF CHICKEN HEMOGLOBIN D | | Descriptor: | PROTEIN (HEMOGLOBIN D), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, E, Oliveira, M.A, Xie, Q, Ernst, S.R, Riggs, A.F, Hackert, M.L. | | Deposit date: | 1998-12-06 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural and functional analysis of the hemoglobin D component from chicken.
J.Biol.Chem., 274, 1999
|
|
1MFI
 
 | | CRYSTAL STRUCTURE OF MACROPHAGE MIGRATION INHIBITORY FACTOR COMPLEXED WITH (E)-2-FLUORO-P-HYDROXYCINNAMATE | | Descriptor: | 2-FLUORO-3-(4-HYDROXYPHENYL)-2E-PROPENEOATE, PROTEIN (MACROPHAGE MIGRATION INHIBITORY FACTOR) | | Authors: | Taylor, A.B, Johnson Jr, W.H, Czerwinski, R.M, Whitman, C.P, Hackert, M.L. | | Deposit date: | 1998-08-12 | | Release date: | 1999-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of macrophage migration inhibitory factor complexed with (E)-2-fluoro-p-hydroxycinnamate at 1.8 A resolution: implications for enzymatic catalysis and inhibition.
Biochemistry, 38, 1999
|
|
1MFF
 
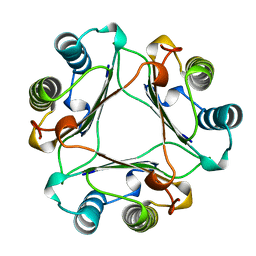 | | MACROPHAGE MIGRATION INHIBITORY FACTOR Y95F MUTANT | | Descriptor: | MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Taylor, A.B, Stamps, S.L, Wang, S.C, Hackert, M.L, Whitman, C.P. | | Deposit date: | 1998-10-19 | | Release date: | 1999-07-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of the phenylpyruvate tautomerase activity of macrophage migration inhibitory factor: properties of the P1G, P1A, Y95F, and N97A mutants.
Biochemistry, 39, 2000
|
|
8LDH
 
 | |
6LDH
 
 | |
1LDM
 
 | |
1ITH
 
 | | STRUCTURE DETERMINATION AND REFINEMENT OF HOMOTETRAMERIC HEMOGLOBIN FROM URECHIS CAUPO AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | CYANIDE ION, HEMOGLOBIN (CYANO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hackert, M, Kolatkar, P, Ernst, S.R, Ogata, C.M, Hendrickson, W.A, Merritt, E.A, Phizackerley, R.P. | | Deposit date: | 1991-12-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination and refinement of homotetrameric hemoglobin from Urechis caupo at 2.5 A resolution.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
