7YWU
 
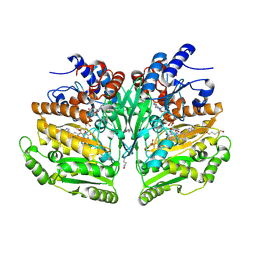 | | Eugenol oxidase from rhodococcus jostii: mutant S81H, A423M, H434Y, S394V, I445D, S518P | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-02-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure- and computational-aided engineering of an oxidase to produce isoeugenol from a lignin-derived compound.
Nat Commun, 13, 2022
|
|
7N5H
 
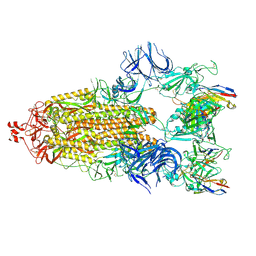 | | Cryo-EM structure of broadly neutralizing antibody 2-36 in complex with prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-36 Fab heavy chain, 2-36 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-03 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | A monoclonal antibody that neutralizes SARS-CoV-2 variants, SARS-CoV, and other sarbecoviruses.
Emerg Microbes Infect, 11, 2022
|
|
7MHS
 
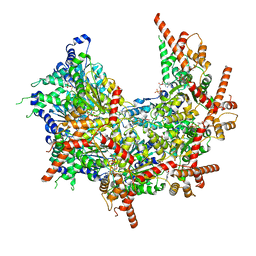 | | Structure of p97 (subunits A to E) with substrate engaged | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Xu, Y, Han, H, Cooney, I, Hill, C.P, Shen, P.S. | | Deposit date: | 2021-04-15 | | Release date: | 2022-05-11 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Active conformation of the p97-p47 unfoldase complex.
Nat Commun, 13, 2022
|
|
7RW2
 
 | |
6UBI
 
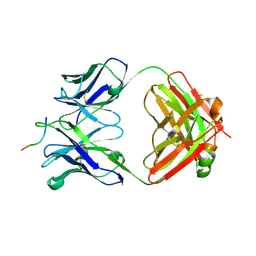 | |
6MQE
 
 | |
6MQS
 
 | |
5JIE
 
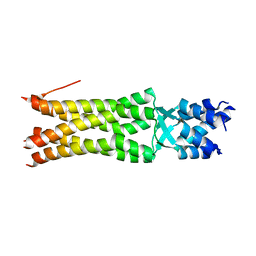 | |
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6NF2
 
 | |
6NZ7
 
 | |
4TKQ
 
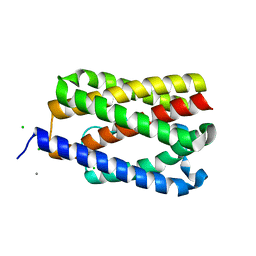 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | Authors: | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8025 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6OT1
 
 | |
6OSY
 
 | |
4WD7
 
 | |
4TKS
 
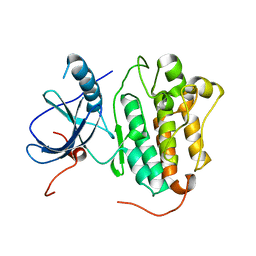 | |
8DS8
 
 | |
6UCE
 
 | |
6UCF
 
 | |
6VIL
 
 | |
7PBG
 
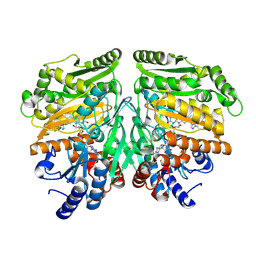 | |
7PBI
 
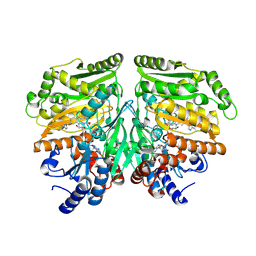 | |
6PZV
 
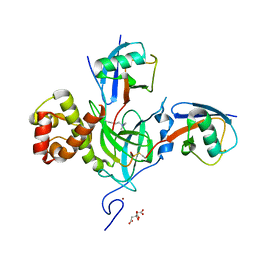 | |
6WXK
 
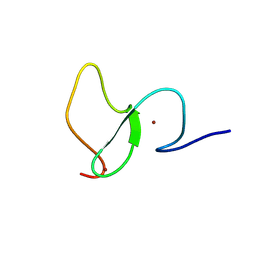 | | PHF23 PHD Domain Apo | | Descriptor: | PHD finger protein 23, ZINC ION | | Authors: | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | Deposit date: | 2020-05-11 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
3PMT
 
 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | Descriptor: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | Authors: | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
