4AHW
 
 | |
4AHY
 
 | | Flo5A cocrystallized with 3 mM GdAc3 | | Descriptor: | CHLORIDE ION, FLOCCULATION PROTEIN FLO5, GADOLINIUM ATOM | | Authors: | Veelders, M, Essen, L.-O. | | Deposit date: | 2012-02-07 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Complex Gadolinium-Oxo Clusters Formed Along Concave Protein Surfaces.
Chembiochem, 13, 2012
|
|
2J09
 
 | | Thermus DNA photolyase with FMN antenna chromophore | | Descriptor: | CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J07
 
 | | Thermus DNA photolyase with 8-HDF antenna chromophore | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J08
 
 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2JGP
 
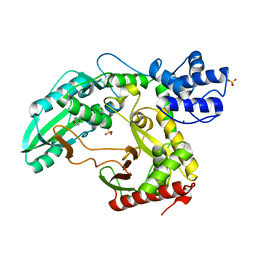 | | Structure of the TycC5-6 PCP-C bidomain of the tyrocidine synthetase TycC | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, SULFATE ION, ... | | Authors: | Samel, S.A, Schoenafinger, G, Knappe, T.A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2007-02-13 | | Release date: | 2007-10-30 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights Into a Peptide Bond-Forming Bidomain from a Nonribosomal Peptide Synthetase.
Structure, 15, 2007
|
|
2JJA
 
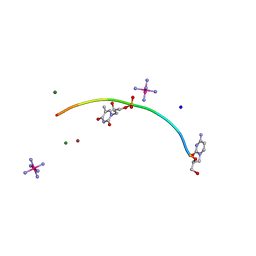 | | Crystal structure of GNA with synthetic copper base pair | | Descriptor: | COBALT HEXAMMINE(III), COPPER (II) ION, GNA, ... | | Authors: | Schlegel, M.K, Essen, L.-O, Meggers, E. | | Deposit date: | 2008-03-28 | | Release date: | 2008-07-22 | | Last modified: | 2022-06-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Duplex Structure of a Minimal Nucleic Acid.
J.Am.Chem.Soc., 130, 2008
|
|
2WNA
 
 | |
2YJ0
 
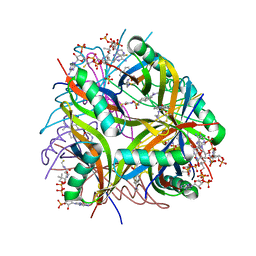 | | X-ray structure of chemically engineered Mycobacterium tuberculosis Dodecin | | Descriptor: | CHLORIDE ION, COENZYME A, DODECIN, ... | | Authors: | Vinzenz, X, Grosse, W, Linne, U, Meissner, B, Essen, L.-O. | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical Engineering of Mycobacterium Tuberculosis Dodecin Hybrids.
Chem.Commun.(Camb.), 47, 2011
|
|
2XJP
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and mannose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJS
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and a1,2-mannobiose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJV
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae with mutation D201T in complex with calcium and glucose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XHG
 
 | | Crystal Structure of the Epimerization Domain from the Initiation Module of Tyrocidine Biosynthesis | | Descriptor: | GLYCEROL, SODIUM ION, TYROCIDINE SYNTHETASE A | | Authors: | Samel, S.-A, Heine, A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2010-06-15 | | Release date: | 2011-07-06 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Epimerization Domain of Tyrocidine Synthetase A
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2Y5C
 
 | | Structure of human ferredoxin 2 (Fdx2)in complex with 2Fe2S cluster | | Descriptor: | ADRENODOXIN-LIKE PROTEIN, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Webert, H, Hobler, A, Sheftel, A.D, Molik, S, Maestre-Reyna, M, Essen, L.-O, Vorniscescu, D, Keusgen, M, Hannemann, F, Bernhardt, R, Lill, R. | | Deposit date: | 2011-01-12 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Functional Studies on Human Mitochondrial Ferredoxins
To be Published
|
|
2XJU
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae with mutation S227A in complex with calcium and a1,2-mannobiose | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XJQ
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, FLOCCULATION PROTEIN FLO5, GLYCEROL, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2YIZ
 
 | | X-ray structure of Mycobacterium tuberculosis Dodecin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Vinzenz, X, Grosse, W, Linne, U, Meissner, B, Essen, L.-O. | | Deposit date: | 2011-05-17 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chemical Engineering of Mycobacterium Tuberculosis Dodecin Hybrids.
Chem.Commun.(Camb.), 47, 2011
|
|
2VTB
 
 | | Structure of cryptochrome 3 - DNA complex | | Descriptor: | 5'-D(*DT*DT*DT*DT*DTP)-3', 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, ACETATE ION, ... | | Authors: | Pokorny, R, Klar, T, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Recognition and Repair of Uv Lesions in Loop Structures of Duplex DNA by Dash-Type Cryptochrome.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7ZHF
 
 | |
7ZHK
 
 | |
4COY
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Galb1-4GlcNAc | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4CP2
 
 | | Crystal Structure of Epithelial Adhesin 9 A domain (Epa9A) from Candida glabrata in complex with Galb1-4GlcNAc | | Descriptor: | CALCIUM ION, CHLORIDE ION, EPITHELIAL ADHESIN 9, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional reprogramming of Candida glabrata epithelial adhesins: the role of conserved and variable structural motifs in ligand binding
To Be Published
|
|
4D3W
 
 | | Crystal Structure of Epithelial Adhesin 1 A domain (Epa1A) from Candida glabrata in complex with the T-antigen (Galb1-3GalNAc) | | Descriptor: | CALCIUM ION, EPITHELIAL ADHESIN 1, GLYCEROL, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4COU
 
 | | Crystal Structure of Epithelial Adhesin 6 A domain (Epa6A) from Candida glabrata in complex with Lactose | | Descriptor: | ACETATE ION, CALCIUM ION, EPITHELIAL ADHESIN 6, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
4CP0
 
 | | Crystal Structure of Epithelial Adhesin 9 A domain (Epa9A) from Candida glabrata in complex with Lactose | | Descriptor: | CALCIUM ION, CHLORIDE ION, EPITHELIAL ADHESIN 9, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional reprogramming of Candida glabrata epithelial adhesins: the role of conserved and variable structural motifs in ligand binding
To Be Published
|
|
