6SLX
 
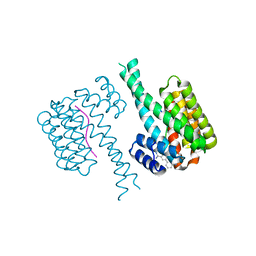 | | Fragment AZ-010 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, TAZpS89, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]propanamide | | Authors: | Ottmann, C, Wolter, M, Guillory, X, Genet, S, Somsen, B, Leysen, S, Castaldi, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.800045 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6SLW
 
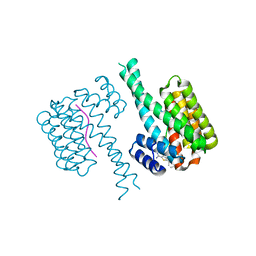 | | Fragment AZ-004 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-methyl-5-phenyl-thiophene-2-carboximidamide, WW domain-containing transcription regulator protein 1 | | Authors: | Ottmann, C, Wolter, M, Guillory, X, Leysen, S, Genet, S, Somsen, B, Patel, J, Castaldi, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6U0D
 
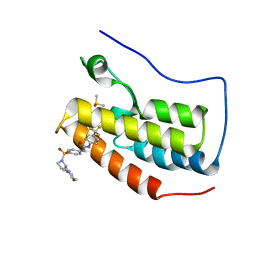 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0590 | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, N-[4-({(2S)-2-[(morpholin-4-yl)methyl]pyrrolidin-1-yl}sulfonyl)phenyl]-N'-[4-(trifluoromethyl)phenyl]urea | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Discovery, X-ray Crystallography, and Anti-inflammatory Activity of Bromodomain-containing Protein 4 (BRD4) BD1 Inhibitors Targeting a Distinct New Binding Site
J. Med. Chem., 2022
|
|
6R5L
 
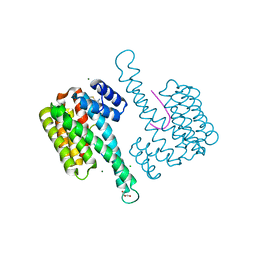 | | Fragment AZ-006 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{S})-1-azanylpropan-2-yl]amino]-6-(sulfanylmethyl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RK8
 
 | | Fragment AZ-014 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(3-azanyl-4-methyl-pyrazol-1-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RL3
 
 | | Fragment AZ-003 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RL6
 
 | | Fragment AZ-024 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
4ZK5
 
 | | MAP4K4 in complex with inhibitor GNE-495 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-amino-N-[1-(cyclopropylcarbonyl)azetidin-3-yl]-2-(3-fluorophenyl)-1,7-naphthyridine-5-carboxamide, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2015-04-29 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure-Based Design of GNE-495, a Potent and Selective MAP4K4 Inhibitor with Efficacy in Retinal Angiogenesis.
Acs Med.Chem.Lett., 6, 2015
|
|
5KE0
 
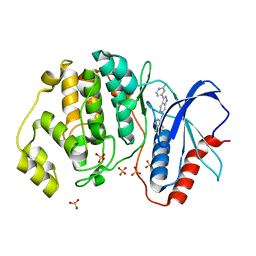 | | Discovery of 1-1H-Pyrazolo 4,3-c pyridine-6-yl urea Inhibitors of Extracellular Signal Regulated Kinase ERK for the Treatment of Cancers | | Descriptor: | 1-[3-(2-methylpyridin-4-yl)-1~{H}-pyrazolo[4,3-c]pyridin-6-yl]-3-(phenylmethyl)urea, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Lim, J. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of 1-(1H-Pyrazolo[4,3-c]pyridin-6-yl)urea Inhibitors of Extracellular Signal-Regulated Kinase (ERK) for the Treatment of Cancers.
J.Med.Chem., 59, 2016
|
|
8DTO
 
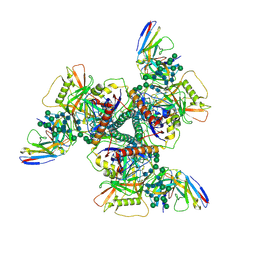 | | Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-07-26 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
8DY6
 
 | | Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stalls, V, Acharya, P. | | Deposit date: | 2022-08-03 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Mutation-guided vaccine design: A process for developing boosting immunogens for HIV broadly neutralizing antibody induction.
Cell Host Microbe, 32, 2024
|
|
5MHC
 
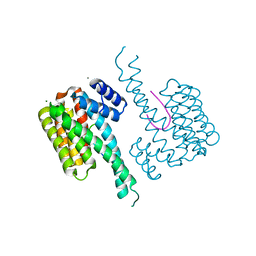 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, LYS-LEU-MET-PHE-LYS-TPO-GLU-GLY-PRO-ASP-SER-ASP, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-11-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
5MOC
 
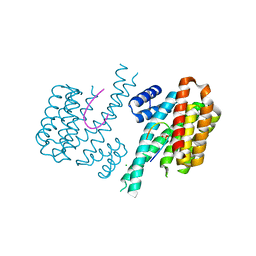 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
5MXO
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide stabilized by Fusicoccin-A | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, FUSICOCCIN, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
8W89
 
 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W87
 
 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8A
 
 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W88
 
 | | Cryo-EM structure of the SEP363856-bound TAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8B
 
 | | Cryo-EM structure of SEP-363856 bounded serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6SMB
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | Tyrosine-protein kinase JAK1, ~{N}-[3-[2-[(3-methoxy-1-methyl-pyrazol-4-yl)amino]-5-methyl-pyrimidin-4-yl]-1~{H}-indol-7-yl]-2-methyl-pyridine-3-carboxamide | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6SM8
 
 | | Human jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-6-[(3~{S})-3-[(1~{S})-2-cyano-1-[4-(7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl)pyrazol-1-yl]ethyl]pyrrolidin-1-yl]benzenecarbonitrile, Tyrosine-protein kinase JAK1 | | Authors: | Read, J.A, Steuber, H. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-29 | | Last modified: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (2R)-N-[3-[2-[(3-Methoxy-1-methyl-pyrazol-4-yl)amino]pyrimidin-4-yl]-1H-indol-7-yl]-2-(4-methylpiperazin-1-yl)propenamide (AZD4205) as a Potent and Selective Janus Kinase 1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
3EQB
 
 | | X-ray structure of the human mitogen-activated protein kinase kinase 1 (MEK1) in a complex with ligand and MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Ohren, J.F, Pavlovsky, A, Zhang, E. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | 2-Alkylamino- and alkoxy-substituted 2-amino-1,3,4-oxadiazoles-O-Alkyl benzohydroxamate esters replacements retain the desired inhibition and selectivity against MEK (MAP ERK kinase).
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6P7I
 
 | | Crystal structure of Human PRMT6 in complex with S-Adenosyl-L-Homocysteine and YS17-117 Compound | | Descriptor: | GLYCEROL, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]prop-2-enamide, N-[3-(4-{[(2-aminoethyl)(methyl)amino]methyl}-1H-pyrrol-3-yl)phenyl]propanamide, ... | | Authors: | Halabelian, L, Dong, A, Zeng, H, Li, Y, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a First-in-Class Protein Arginine Methyltransferase 6 (PRMT6) Covalent Inhibitor
J.Med.Chem., 63, 2020
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
6CZS
 
 | |
