1SFV
 
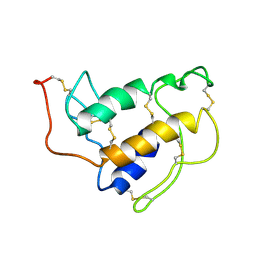 | | PORCINE PANCREAS PHOSPHOLIPASE A2, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Van Den Berg, B, Tessari, M, Boelens, R, Dijkman, R, Kaptein, R, De Haas, G.H, Verheij, H.M. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic phospholipase A2 complexed with micelles and a competitive inhibitor.
J.Biomol.NMR, 5, 1995
|
|
2C06
 
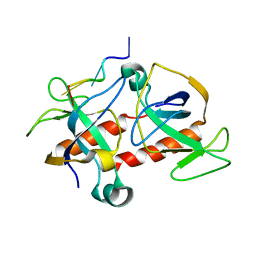 | | NMR-based model of the complex of the toxin Kid and a 5-nucleotide substrate RNA fragment (AUACA) | | Descriptor: | 5'-R(*AP*UP*AP*CP*AP)-3', KID TOXIN PROTEIN | | Authors: | Kamphuis, M.B, Bonvin, A.M.J.J, Monti, M.C, Lemonnier, M, Munoz-Gomez, A, Van Den Heuvel, R.H.H, Diaz-Orejas, R, Boelens, R. | | Deposit date: | 2005-08-25 | | Release date: | 2006-02-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Model for RNA Binding and the Catalytic Site of the Rnase Kid of the Bacterial Pard Toxin-Antitoxin System.
J.Mol.Biol., 357, 2006
|
|
1QEY
 
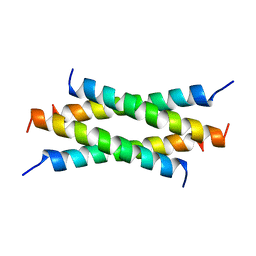 | | NMR Structure Determination of the Tetramerization Domain of the MNT Repressor: An Asymmetric A-Helical Assembly in Slow Exchange | | Descriptor: | PROTEIN (REGULATORY PROTEIN MNT) | | Authors: | Nooren, I.M.A, George, A.V.E, Kaptein, R, Sauer, R.T, Boelens, R. | | Deposit date: | 1999-04-03 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The tetramerization domain of the Mnt repressor consists of two right-handed coiled coils.
Nat.Struct.Biol., 6, 1999
|
|
1RGD
 
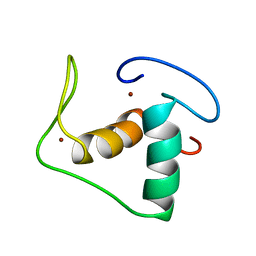 | | STRUCTURE REFINEMENT OF THE GLUCOCORTICOID RECEPTOR-DNA BINDING DOMAIN FROM NMR DATA BY RELAXATION MATRIX CALCULATIONS | | Descriptor: | GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Van Tilborg, M.A.A, Bonvin, A.M.J.J, Hard, K, Davis, A, Maler, B, Boelens, R, Yamamoto, K.R, Kaptein, R. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure refinement of the glucocorticoid receptor-DNA binding domain from NMR data by relaxation matrix calculations.
J.Mol.Biol., 247, 1995
|
|
1AH9
 
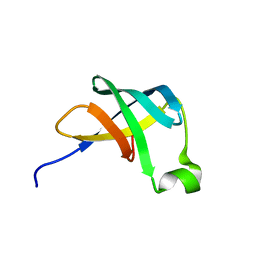 | | THE STRUCTURE OF THE TRANSLATIONAL INITIATION FACTOR IF1 FROM ESCHERICHIA COLI, NMR, 19 STRUCTURES | | Descriptor: | INITIATION FACTOR 1 | | Authors: | Sette, M, Van Tilborg, P, Spurio, R, Kaptein, R, Paci, M, Gualerzi, C.O, Boelens, R. | | Deposit date: | 1997-04-16 | | Release date: | 1997-07-07 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif.
EMBO J., 16, 1997
|
|
1L1M
 
 | | SOLUTION STRUCTURE OF A DIMER OF LAC REPRESSOR DNA-BINDING DOMAIN COMPLEXED TO ITS NATURAL OPERATOR O1 | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3', 5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3', Lactose operon repressor | | Authors: | Kalodimos, C.G, Bonvin, A.M.J.J, Salinas, R.K, Wechselberger, R, Boelens, R, Kaptein, R. | | Deposit date: | 2002-02-19 | | Release date: | 2002-06-26 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Plasticity in protein-DNA recognition: lac repressor interacts with its natural operator 01 through alternative conformations of its DNA-binding domain.
EMBO J., 21, 2002
|
|
1HD4
 
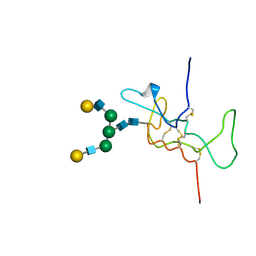 | | SOLUTION STRUCTURE OF THE A-SUBUNIT OF HUMAN CHORIONIC GONADOTROPIN [MODELED WITH DIANTENNARY GLYCAN AT ASN78] | | Descriptor: | CHORIONIC GONADOTROPIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Erbel, P.J.A, Karimi-Nejad, Y, van Kuik, J.A, Boelens, R, Kamerling, J.P, Vliegenthart, J.F.G. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-07 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Effects of the N-Linked Glycans on the 3D Structure of the Free A-Subunit of Human Chorionic Gonadotropin
Biochemistry, 39, 2000
|
|
1QMC
 
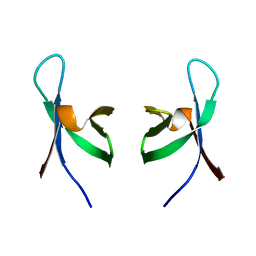 | | C-terminal DNA-binding domain of HIV-1 integrase, NMR, 42 structures | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Eijkelenboom, A.P.A.M, Sprangers, R, Hard, K, Puras Lutzke, R.A, Plasterk, R.H.A, Boelens, R, Kaptein, R. | | Deposit date: | 1999-09-27 | | Release date: | 1999-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Refined Solution Structure of the C-Terminal DNA-Binding Domain of Human Immunovirus-1 Integrase.
Proteins: Struct.,Funct., Genet., 36, 1999
|
|
2YEN
 
 | | Solution structure of the skeletal muscle and neuronal voltage gated sodium channel antagonist mu-conotoxin CnIIIC | | Descriptor: | Mu-conotoxin CnIIIC | | Authors: | Favreau, P, Benoit, E, Hocking, H.G, Carlier, L, D'hoedt, D, Leipold, E, Markgraf, R, Schlumberger, S, Cordova, M.A, Gaertner, H, Paolini-Bertrand, M, Hartley, O, Tytgat, J, Heinemann, S.H, Bertrand, D, Boelens, R, Stocklin, R, Molgo, J. | | Deposit date: | 2011-03-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Novel Mu-Conopeptide, Cniiic, Exerts Potent and Preferential Inhibition of Na(V) 1.2/1.4 Channels and Blocks Neuronal Nicotinic Acetylcholine Receptors.
Br.J.Pharmacol., 166, 2012
|
|
1B28
 
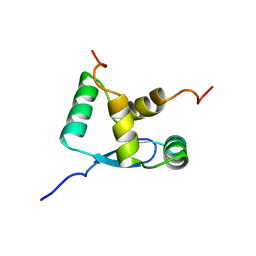 | | ARC REPRESSOR MYL MUTANT FROM SALMONELLA BACTERIOPHAGE P22 | | Descriptor: | PROTEIN (REGULATORY PROTEIN ARC) | | Authors: | Rietveld, A.W.M, Nooren, I.M.A, Sauer, R.T, Kaptein, R, Boelens, R. | | Deposit date: | 1998-12-05 | | Release date: | 1999-11-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of an Arc repressor mutant reveal premelting conformational changes related to DNA binding.
Biochemistry, 38, 1999
|
|
1BRV
 
 | | SOLUTION NMR STRUCTURE OF THE IMMUNODOMINANT REGION OF PROTEIN G OF BOVINE RESPIRATORY SYNCYTIAL VIRUS, 48 STRUCTURES | | Descriptor: | PROTEIN G | | Authors: | Doreleijers, J.F, Langedijk, J.P.M, Hard, K, Rullmann, J.A.C, Boelens, R, Schaaper, W.M, Van Oirschot, J.T, Kaptein, R. | | Deposit date: | 1996-03-29 | | Release date: | 1997-06-05 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant region of protein G of bovine respiratory syncytial virus.
Biochemistry, 35, 1996
|
|
2BUN
 
 | | Solution structure of the BLUF domain of AppA 5-125 | | Descriptor: | APPA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Grinstead, J.S, Hsu, S.-T, Laan, W, Bonvin, A.M.J.J, Hellingwerf, K.J, Boelens, R, Kaptein, R. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling.
Chembiochem, 7, 2006
|
|
1XFQ
 
 | | structure of the blue shifted intermediate state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1XFN
 
 | | NMR structure of the ground state of the photoactive yellow protein lacking the N-terminal part | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Bernard, C, Houben, K, Derix, N.M, Marks, D, van der Horst, M.A, Hellingwerf, K.J, Boelens, R, Kaptein, R, van Nuland, N.A. | | Deposit date: | 2004-09-15 | | Release date: | 2005-08-16 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a transient photoreceptor intermediate: delta25 photoactive yellow protein
STRUCTURE, 13, 2005
|
|
1M1F
 
 | | Kid toxin protein from E.coli plasmid R1 | | Descriptor: | Kid toxin protein, PHOSPHATE ION | | Authors: | Hargreaves, D, Santos-Sierra, S, Giraldo, R, Sabariegos-Jareno, R, de la Cueva-Mendez, G, Boelens, R, Diaz-Orejas, R, Rafferty, J.B. | | Deposit date: | 2002-06-19 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Analysis of the Kid toxin protein from E.coli plasmid R1
Structure, 10, 2002
|
|
1Z00
 
 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
1Z9B
 
 | | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2 | | Descriptor: | Translation initiation factor IF-2 | | Authors: | Wienk, H, Tomaselli, S, Bernard, C, Spurio, R, Picone, D, Gualerzi, C.O, Boelens, R. | | Deposit date: | 2005-04-01 | | Release date: | 2005-08-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C1-subdomain of Bacillus stearothermophilus translation initiation factor IF2
Protein Sci., 14, 2005
|
|
2AQ0
 
 | | Solution structure of the human homodimeric dna repair protein XPF | | Descriptor: | DNA repair endonuclease XPF | | Authors: | Das, D, Tripsianes, K, Folkers, G, Jaspers, N.G, Hoeijmakers, J.H, Kaptein, R, Boelens, R. | | Deposit date: | 2005-08-17 | | Release date: | 2006-10-03 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The HhH domain of the human DNA repair protein XPF forms stable homodimers
Proteins, 70, 2008
|
|
1KFT
 
 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
1T3O
 
 | | Solution structure of CsrA, a bacterial carbon storage regulatory protein | | Descriptor: | Carbon storage regulator | | Authors: | Koharudin, L.M.I, Georgiou, T, Kleanthous, C, Geoffrey, R, Kaptein, R, Boelens, R. | | Deposit date: | 2004-04-27 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A model for RNA binding by the bacterial carbon storage regulatory protein, CsrA
To be Published
|
|
1PCO
 
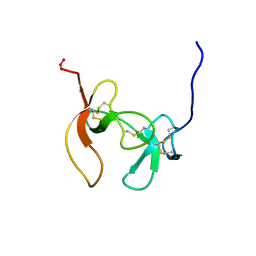 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | HYDROXIDE ION, PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
1PCN
 
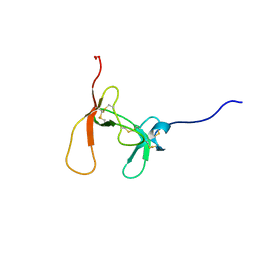 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | HYDROXIDE ION, PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
1POG
 
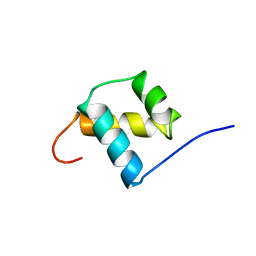 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
1LEB
 
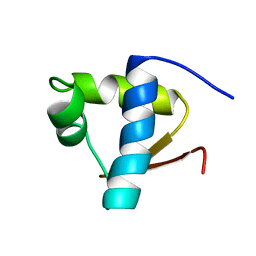 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
1LEA
 
 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
