1TIP
 
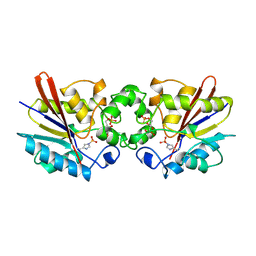 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, PHOSPHOENZYME INTERMEDIATE OF FRU-2,6-BISPHOSPHATASE | | Authors: | Lee, Y.-H, Olson, T.W, Ogata, C.M, Levitt, D.G, Banaszak, L.J, Lange, A.J. | | Deposit date: | 1997-05-28 | | Release date: | 1998-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
Nat.Struct.Biol., 4, 1997
|
|
1EMD
 
 | |
1FBT
 
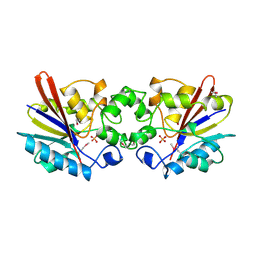 | | THE BISPHOSPHATASE DOMAIN OF THE BIFUNCTIONAL RAT LIVER 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE | | Descriptor: | FRUCTOSE-2,6-BISPHOSPHATASE, PHOSPHATE ION | | Authors: | Lee, Y.-H, Ogata, C, Pflugrath, J.W, Levitt, D.G, Sarma, R, Banaszak, L.J, Pilkis, S.J. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the rat liver fructose-2,6-bisphosphatase based on selenomethionine multiwavelength anomalous dispersion phases.
Biochemistry, 35, 1996
|
|
1G74
 
 | |
1G7N
 
 | |
1IL0
 
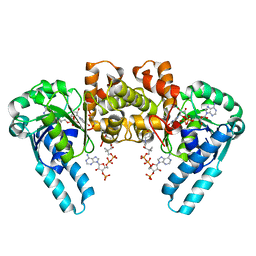 | | X-RAY CRYSTAL STRUCTURE OF THE E170Q MUTANT OF HUMAN L-3-HYDROXYACYL-COA DEHYDROGENASE | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, ACETOACETYL-COENZYME A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2001-05-07 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glutamate 170 of human l-3-hydroxyacyl-CoA dehydrogenase is required for proper orientation of the catalytic histidine and structural integrity of the enzyme.
J.Biol.Chem., 276, 2001
|
|
1LIF
 
 | |
1LIB
 
 | |
1LSO
 
 | |
1LID
 
 | |
1LSJ
 
 | |
1LSH
 
 | |
1M75
 
 | |
1M76
 
 | |
1HQS
 
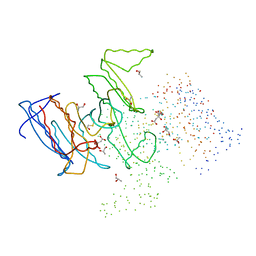 | | CRYSTAL STRUCTURE OF ISOCITRATE DEHYDROGENASE FROM BACILLUS SUBTILIS | | Descriptor: | CITRIC ACID, ISOCITRATE DEHYDROGENASE, R-1,2-PROPANEDIOL, ... | | Authors: | Singh, S.K, Matsuno, K, LaPorte, D.C, Banaszak, L.J. | | Deposit date: | 2000-12-19 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase.
J.Biol.Chem., 276, 2001
|
|
1IB6
 
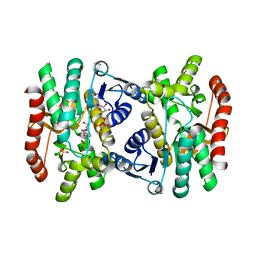 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-03-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1IE3
 
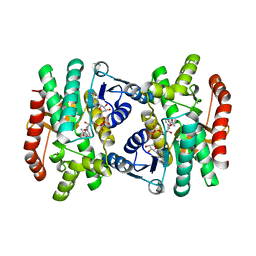 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1IFB
 
 | |
1LIE
 
 | |
1LIC
 
 | |
5MDH
 
 | | CRYSTAL STRUCTURE OF TERNARY COMPLEX OF PORCINE CYTOPLASMIC MALATE DEHYDROGENASE ALPHA-KETOMALONATE AND TNAD AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-KETOMALONIC ACID, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chapman, A.D.M, Cortes, A, Dafforn, T.R, Clarke, A.R, Brady, R.L. | | Deposit date: | 1998-10-08 | | Release date: | 1999-05-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, alpha-ketomalonate and tetrahydoNAD.
J.Mol.Biol., 285, 1999
|
|
1BMD
 
 | |
1A2D
 
 | | PYRIDOXAMINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN, CHLORIDE ION | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-29 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
1AB0
 
 | | C1G/V32D/F57H MUTANT OF MURINE ADIPOCYTE LIPID BINDING PROTEIN AT PH 4.5 | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Kane, C, Simpson, M, Banaszak, L, Bernlohr, D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
J.Biol.Chem., 272, 1997
|
|
1A18
 
 | | PHENANTHROLINE MODIFIED MURINE ADIPOCYTE LIPID BINDING PROTEIN | | Descriptor: | ADIPOCYTE LIPID BINDING PROTEIN | | Authors: | Ory, J, Mazhary, A, Kuang, H, Davies, R, Distefano, M, Banaszak, L. | | Deposit date: | 1997-12-23 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of two synthetic catalysts based on adipocyte lipid-binding protein.
Protein Eng., 11, 1998
|
|
