9MSI
 
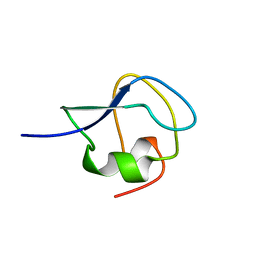 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18N | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
9AME
 
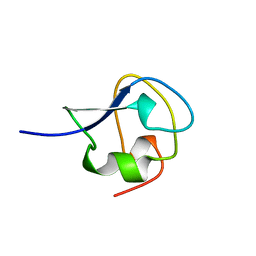 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 S42G | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
8MSI
 
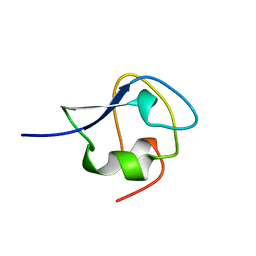 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14SQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
4MSI
 
 | |
6H2B
 
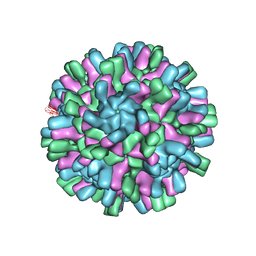 | | Structure of the Macrobrachium rosenbergii Nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
3AME
 
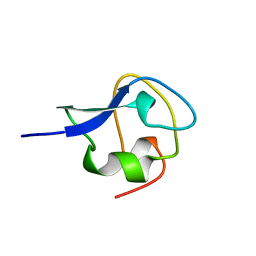 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 Q9TQ44T | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1MSJ
 
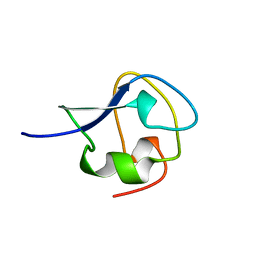 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
4WCJ
 
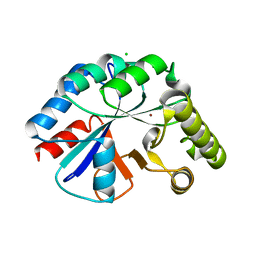 | | Structure of IcaB from Ammonifex degensii | | Descriptor: | CHLORIDE ION, Polysaccharide deacetylase, ZINC ION | | Authors: | Little, D.J, Bamford, N.C, Pokrovskaya, V, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2014-09-04 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the De-N-acetylation of Poly-beta-1,6-N-acetyl-d-glucosamine in Gram-positive Bacteria.
J.Biol.Chem., 289, 2014
|
|
4WCX
 
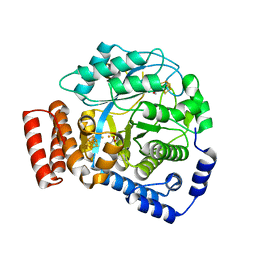 | | Crystal structure of HydG: A maturase of the [FeFe]-hydrogenase | | Descriptor: | ALANINE, Biotin and thiamin synthesis associated, FE (III) ION, ... | | Authors: | Dinis, P.C, Harmer, J.E, Driesener, R.C, Roach, P.L. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray crystallographic and EPR spectroscopic analysis of HydG, a maturase in [FeFe]-hydrogenase H-cluster assembly.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6R62
 
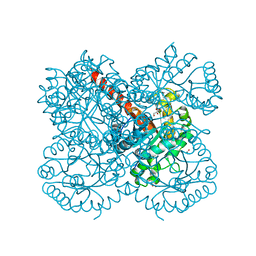 | | Crystal structure of a class II pyruvate aldolase from Sphingomonas wittichii RW1 in complex with hydroxypyruvate | | Descriptor: | 3-HYDROXYPYRUVIC ACID, ACETATE ION, BROMIDE ION, ... | | Authors: | Marsden, S.R, Mestrom, L, Hagedoorn, P.L, Bento, I, McMillan, D.G.G, Hanefeld, U. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | CH-Pi Interactions Promote the Conversion of Hydroxypyruvate in a Class II Pyruvate Aldolase
Adv.Synth.Catal., 2019
|
|
7T8N
 
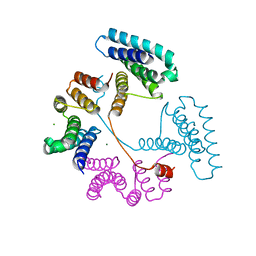 | | Crystal structure of the PNAG binding module PgaA-TPR 220-359 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Poly-beta-1,6-N-acetyl-D-glucosamine export protein | | Authors: | Pfoh, R, Little, D.J, Howell, P.L. | | Deposit date: | 2021-12-16 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The TPR domain of PgaA is a multifunctional scaffold that binds PNAG and modulates PgaB-dependent polymer processing.
Plos Pathog., 18, 2022
|
|
3ULT
 
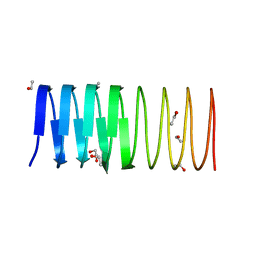 | | Crystal structure of an ice-binding protein from the perennial ryegrass, Lolium perenne | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, Ice recrystallization inhibition protein-like protein | | Authors: | Middleton, A.J, Faucher, F, Campbell, R.L, Davies, P.L. | | Deposit date: | 2011-11-11 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antifreeze protein from freeze-tolerant grass has a beta-roll fold with an irregularly structured ice-binding site.
J.Mol.Biol., 416, 2012
|
|
2G8J
 
 | | Calpain 1 proteolytic core in complex with SNJ-1945, a alpha-ketoamide-type inhibitor. | | Descriptor: | ((1S)-1-((((1S)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2,3-DIOXOPROPYL)AMINO)CARBONYL)-3-METHYLBUTYL)CARBAMIC ACID 5-METHOXY-3-OXAPENTYL ESTER, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
2G8E
 
 | | Calpain 1 proteolytic core in complex with SNJ-1715, a cyclic hemiacetal-type inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Calpain-1 catalytic subunit, ... | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
1MDW
 
 | | Crystal Structure of Calcium-Bound Protease Core of Calpain II Reveals the Basis for Intrinsic Inactivation | | Descriptor: | CALCIUM ION, Calpain II, catalytic subunit | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Jia, Z, Davies, P.L. | | Deposit date: | 2002-08-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Calpain silencing by a reversible intrinsic mechanism.
Nat.Struct.Biol., 10, 2003
|
|
4AME
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
8OIU
 
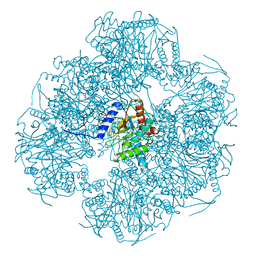 | | Cryo-EM reconstruction of the native 24-mer E2o core of the 2-oxoglutarate dehydrogenase complex of C. thermophilum at 3.35 A resolution | | Descriptor: | Dihydrolipoyllysine-residue succinyltransferase | | Authors: | Skalidis, I, Tueting, C, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2023-03-23 | | Release date: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of an endogenous 4-megadalton succinyl-CoA-generating metabolon.
Commun Biol, 6, 2023
|
|
8PE8
 
 | |
8FT8
 
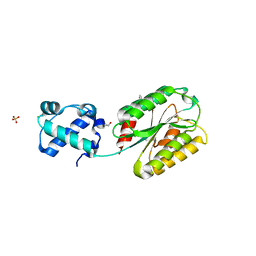 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Thr-Val linker variant, SUMO tag-free preparation | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-11 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FZU
 
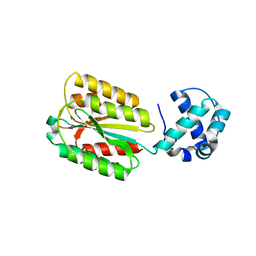 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Thr-Val linker variant, Expressed with SUMO tag | | Descriptor: | POTASSIUM ION, SULFATE ION, Transcription factor ETV6,Anthrax toxin receptor 2 | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Lebedev, A, Moody, J.D. | | Deposit date: | 2023-01-30 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8FT6
 
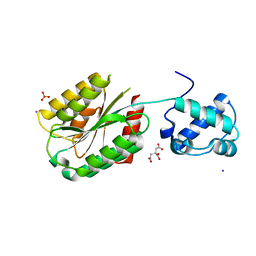 | | The von Willebrand factor A domain of human capillary morphogenesis gene II, flexibly fused to the 1TEL crystallization chaperone, Ala-Ala linker variant, SUMO tag-free preparation. | | Descriptor: | CITRIC ACID, IODIDE ION, SULFATE ION, ... | | Authors: | Gajjar, P.L, Litchfield, C.M, Callahan, M, Redd, N, Doukov, T, Moody, J.D. | | Deposit date: | 2023-01-11 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8AME
 
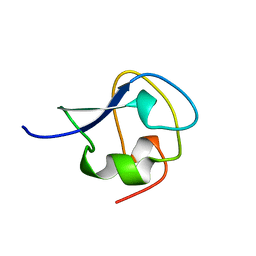 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 N14SA16H | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
7AME
 
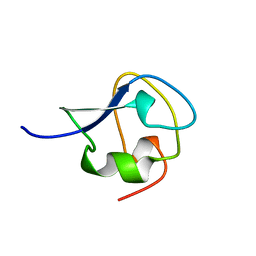 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15A | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1N4I
 
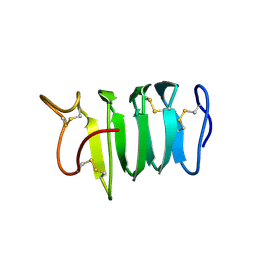 | | Solution structure of spruce budworm antifreeze protein at 5 degrees celsius | | Descriptor: | thermal hysteresis protein | | Authors: | Graether, S.P, Gagne, S.M, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-10-31 | | Release date: | 2003-04-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Spruce Budworm Antifreeze Protein: Changes in Structure and Dynamics at Low Temperature
J.Mol.Biol., 327, 2003
|
|
6SHT
 
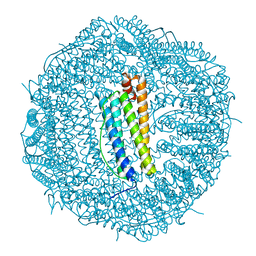 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | Descriptor: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | Authors: | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | Deposit date: | 2019-08-08 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
