1PWT
 
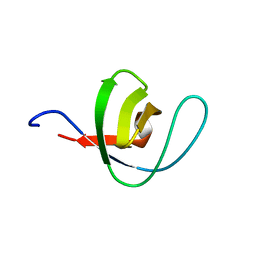 | | THERMODYNAMIC ANALYSIS OF ALPHA-SPECTRIN SH3 AND TWO OF ITS CIRCULAR PERMUTANTS WITH DIFFERENT LOOP LENGTHS: DISCERNING THE REASONS FOR RAPID FOLDING IN PROTEINS | | Descriptor: | ALPHA SPECTRIN | | Authors: | Martinez, J.C, Viguera, A.R, Berisio, R, Wilmanns, M, Mateo, P.L, Filmonov, V.V, Serrano, L. | | Deposit date: | 1998-10-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermodynamic analysis of alpha-spectrin SH3 and two of its circular permutants with different loop lengths: discerning the reasons for rapid folding in proteins.
Biochemistry, 38, 1999
|
|
1BK2
 
 | |
2F0R
 
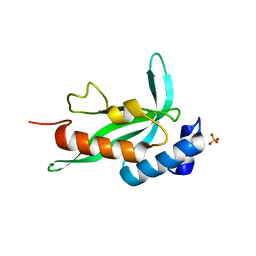 | | Crystallographic structure of human Tsg101 UEV domain | | Descriptor: | SULFATE ION, Tumor susceptibility gene 101 protein | | Authors: | Camara-Artigas, A, Luque, I, Palencia, A, Martinez, J.C, Mateo, P.L. | | Deposit date: | 2005-11-13 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of human TSG101 UEV domain.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2JMC
 
 | | Chimer between Spc-SH3 and P41 | | Descriptor: | Spectrin alpha chain, brain and P41 peptide chimera | | Authors: | van Nuland, N.A.J, Candel, A.M, Martinez, J.C, Conejero-Lara, F, Bruix, M. | | Deposit date: | 2006-11-02 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The high-resolution NMR structure of a single-chain chimeric protein mimicking a SH3-peptide complex
Febs Lett., 581, 2007
|
|
6XVN
 
 | |
6XVM
 
 | |
6XVO
 
 | |
1QKX
 
 | |
1QKW
 
 | | Alpha-spectrin Src Homology 3 domain, N47G mutant in the distal loop. | | Descriptor: | ALPHA II SPECTRIN, GLYCEROL, SULFATE ION | | Authors: | Vega, M.C, Martinez, J, Serrano, L. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural characterization of Asn and Ala residues in the disallowed II' region of the Ramachandran plot.
Protein Sci., 9, 2000
|
|
6XX4
 
 | |
6XX3
 
 | |
6XX5
 
 | |
6XX2
 
 | |
3EG2
 
 | | Crystal structure of the N114Q mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG1
 
 | |
3EGU
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1, SULFATE ION | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG0
 
 | | Crystal structure of the N114T mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
3EG3
 
 | | Crystal structure of the N114A mutant of ABL-SH3 domain | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Camara-Artigas, A. | | Deposit date: | 2008-09-10 | | Release date: | 2009-09-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of interfacial water molecules in proline-rich ligand recognition by the Src homology 3 domain of Abl.
J.Biol.Chem., 285, 2010
|
|
7ZX2
 
 | | Tubulin-Pelophen B complex | | Descriptor: | (3R,4S,7S,9S,11S)-3,4,11-trihydroxy-7-((R,Z)-4-(hydroxymethyl)hex-2-en-2-yl)-9-methoxy-12,12-dimethyl-6-oxa-1(1,3)-benzenacyclododecaphan-5-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Estevez-Gallego, J, Diaz, J.F, Van der Eycken, J, Oliva, M.A. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
8A0L
 
 | | Tubulin-CW1-complex | | Descriptor: | (3~{S},4~{R},8~{S},10~{S},12~{S},14~{S})-14-[(~{Z},4~{R})-4-(hydroxymethyl)hex-2-en-2-yl]-4,12-dimethoxy-9,9-dimethyl-3,8,10-tris(oxidanyl)-1-oxacyclotetradecan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Diaz, J.F, Steinmetz, M.O, Oliva, M.A. | | Deposit date: | 2022-05-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9981 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
8AH5
 
 | |
8AH4
 
 | |
8AH8
 
 | |
8AH6
 
 | |
8AH7
 
 | |
