3WR7
 
 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
3WNI
 
 | |
3X1G
 
 | |
3X1E
 
 | |
3X1F
 
 | |
3X1N
 
 | |
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1WNS
 
 | | Crystal structure of family B DNA polymerase from hyperthermophilic archaeon pyrococcus kodakaraensis KOD1 | | Descriptor: | DNA POLYMERASE | | Authors: | Hashimoto, H, Inoue, T, Kai, Y, Fujiwara, S, Takagi, M, Nishioka, M, Imanaka, T. | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-17 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of DNA Polymerase from Hyperthermophilic Archaeon Pyrococcus Kodakaraensis Kod1
J.Mol.Biol., 306, 2001
|
|
2E2G
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (pre-oxidation form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E2M
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfinic acid form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-14 | | Release date: | 2007-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2CZR
 
 | | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA | | Descriptor: | GLYCEROL, TBP-interacting protein, ZINC ION | | Authors: | Yamamoto, T, Matsuda, T, Inoue, T, Matsumura, H, Morikawa, M, Kanaya, S, Kai, Y. | | Deposit date: | 2005-07-15 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA
Protein Sci., 15, 2006
|
|
2CW7
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2CW8
 
 | | Crystal structure of intein homing endonuclease II | | Descriptor: | Endonuclease PI-PkoII, GLYCEROL, SULFATE ION | | Authors: | Matsumura, H, Takahashi, H, Inoue, T, Hashimoto, H, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-04-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1
Proteins, 63, 2006
|
|
2DV6
 
 | | Crystal structure of nitrite reductase from Hyphomicrobium denitrificans | | Descriptor: | COPPER (II) ION, Nitrite reductase, POTASSIUM ION | | Authors: | Nojiri, M, Xie, Y, Yamamoto, T, Inoue, T, Suzuki, S, Kai, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a hexameric copper-containing nitrite reductase.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1WS8
 
 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (II) ION, GLYCEROL, mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kunishige, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
1WS7
 
 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (I) ION, Mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kataoka, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
2CVD
 
 | | Crystal structure analysis of human hematopoietic prostaglandin D synthase complexed with HQL-79 | | Descriptor: | 4-(BENZHYDRYLOXY)-1-[3-(1H-TETRAAZOL-5-YL)PROPYL]PIPERIDINE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Aritake, K, Kado, Y, Inoue, T, Miyano, M, Urade, Y. | | Deposit date: | 2005-06-02 | | Release date: | 2006-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of HQL-79, an Orally Selective Inhibitor of Human Hematopoietic Prostaglandin D Synthase.
J.Biol.Chem., 281, 2006
|
|
4W4Q
 
 | | Glucose isomerase structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Nango, E, Tanaka, T, Sugahara, M, Suzuki, M, Iwata, S. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat.Methods, 12, 2015
|
|
1GU5
 
 | |
1GTW
 
 | |
1GU4
 
 | |
1H89
 
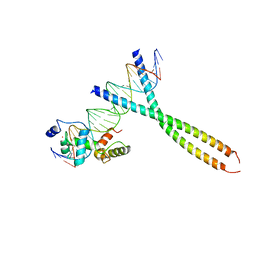 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX2 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-30 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1H8A
 
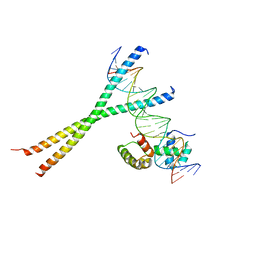 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-31 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
5NEN
 
 | |
1H88
 
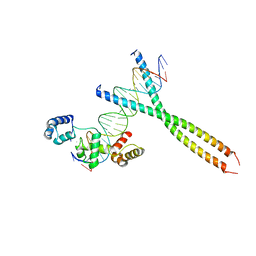 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX1 | | Descriptor: | AMMONIUM ION, CCAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-29 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
