3GEM
 
 | | Crystal structure of short-chain dehydrogenase from Pseudomonas syringae | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Nocek, B, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray crystal structure of short-chain dehydrogenase from Pseudomonas syringae.
To be Published
|
|
3H0X
 
 | | Crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae | | Descriptor: | 78 kDa glucose-regulated protein homolog | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae.
To be Published
|
|
3GO9
 
 | | Predicted insulinase family protease from Yersinia pestis | | Descriptor: | DI(HYDROXYETHYL)ETHER, ZINC ION, insulinase family protease | | Authors: | Osipiuk, J, Maltseva, N, Onopriyenko, O, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-18 | | Release date: | 2009-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structure of predicted insulinase family protease from Yersinia pestis.
To be Published
|
|
3GL1
 
 | | Crystal structure of ATPase domain of Ssb1 chaperone, a member of the HSP70 family, from Saccharomyces cerevisiae | | Descriptor: | CHLORIDE ION, GLYCEROL, Heat shock protein SSB1, ... | | Authors: | Osipiuk, J, Li, H, Bargassa, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of ATPase domain of Ssb1 chaperone, member of the HSP70 family from Saccharomyces cerevisiae.
To be Published
|
|
3HE1
 
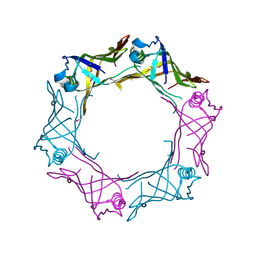 | | Secreted protein Hcp3 from Pseudomonas aeruginosa. | | Descriptor: | GLYCEROL, Major exported Hcp3 protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of secretory protein Hcp3 from Pseudomonas aeruginosa.
J.Struct.Funct.Genom., 12, 2011
|
|
3IC5
 
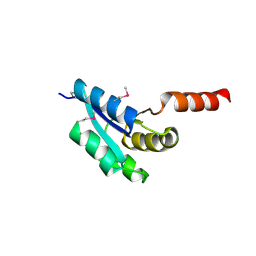 | |
3IR9
 
 | |
3IWG
 
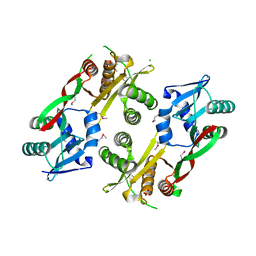 | | Acetyltransferase from GNAT family from Colwellia psychrerythraea. | | Descriptor: | Acetyltransferase, GNAT family, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Stein, A, Shackelford, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of Acetyltransferase from GNAT family from Colwellia psychrerythraea.
To be Published
|
|
3JTJ
 
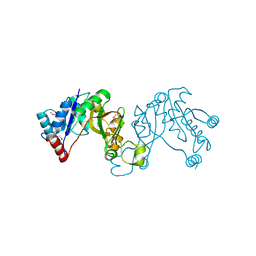 | | 3-deoxy-manno-octulosonate cytidylyltransferase from Yersinia pestis | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase, IMIDAZOLE | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-12 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | X-ray crystal structure of 3-deoxy-manno-octulosonate cytidylyltransferase from Yersinia pestis.
To be Published
|
|
3JUW
 
 | |
3HP7
 
 | | Putative hemolysin from Streptococcus thermophilus. | | Descriptor: | ETHANOL, Hemolysin, putative | | Authors: | Osipiuk, J, Hatzos, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray crystal structure of putative hemolysin from Streptococcus thermophilus.
To be Published
|
|
3I1I
 
 | | X-ray crystal structure of homoserine O-acetyltransferase from Bacillus anthracis | | Descriptor: | ACETATE ION, GLYCEROL, Homoserine O-acetyltransferase, ... | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | X-ray crystal structure of homoserine O-acetyltransferase from Bacillus anthracis.
To be published
|
|
3K1U
 
 | | Beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-xylosidase, ... | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-06 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of
beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum at 1.55 A resolution
To be Published
|
|
3Q7I
 
 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with 6-phosphogluconic acid. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-PHOSPHOGLUCONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3RNS
 
 | |
3NJH
 
 | | D37A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | CALCIUM ION, GLYCEROL, Peptidase, ... | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3Q88
 
 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with ribose 1,5-bisphosphate. | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3RHI
 
 | | DNA-binding protein HU from Bacillus anthracis | | Descriptor: | DNA-binding protein HU | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | DNA-binding protein HU from Bacillus anthracis.
To be Published
|
|
4PV4
 
 | | Proline aminopeptidase P II from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, HEXAETHYLENE GLYCOL, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-03-14 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Proline aminopeptidase P II from Yersinia pestis
To be Published
|
|
4PW0
 
 | |
4Q88
 
 | | Glycosyl hydrolase family 88 from Bacteroides vulgatus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Glycosyl hydrolase Family 88 from Bacteroides vulgatus
To be Published
|
|
4R9N
 
 | |
4QQ3
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP | | Descriptor: | CHLORIDE ION, Inosine-5'-monophosphate dehydrogenase, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from vibrio cholerae, deletion mutant, in complex with xmp
To be Published
|
|
4QNE
 
 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with NAD and IMP | | Descriptor: | INOSINIC ACID, Inosine 5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with NAD and IMP
To be Published
|
|
4S1P
 
 | | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens | | Descriptor: | UNKNOWN LIGAND, Uncharacterized protein | | Authors: | Osipiuk, J, Cuff, M.E, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens
To be Published
|
|
