6Z00
 
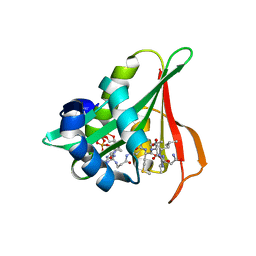 | | Arabidopsis thaliana Naa50 in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA-LEU | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6YZZ
 
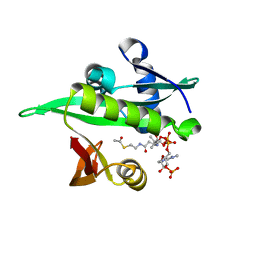 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6ZQQ
 
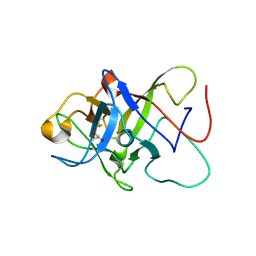 | | Structure of the Pmt3-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT3 isoform 1 | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
6ZQP
 
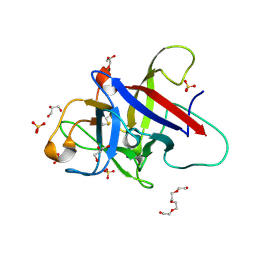 | | Structure of the Pmt2-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT2 isoform 1, SULFATE ION, ... | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
6ZMP
 
 | |
2FH5
 
 | | The Structure of the Mammalian SRP Receptor | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor alpha subunit, ... | | Authors: | Schlenker, O, Wild, K, Sinning, I. | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains.
J.Biol.Chem., 281, 2006
|
|
3SJB
 
 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJC
 
 | |
3SJA
 
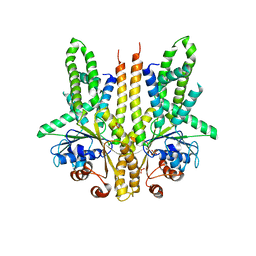 | | Crystal structure of S. cerevisiae Get3 in the open state in complex with Get1 cytosolic domain | | Descriptor: | ATPase GET3, Golgi to ER traffic protein 1, PHOSPHATE ION, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
3SJD
 
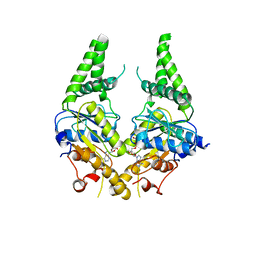 | | Crystal structure of S. cerevisiae Get3 with bound ADP-Mg2+ in complex with Get2 cytosolic domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase GET3, Golgi to ER traffic protein 2, ... | | Authors: | Reitz, S, Wild, K, Sinning, I. | | Deposit date: | 2011-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex.
Science, 333, 2011
|
|
4ZN4
 
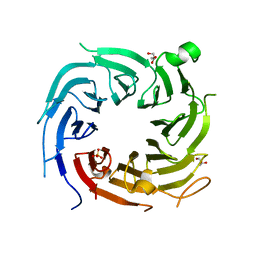 | |
2PX3
 
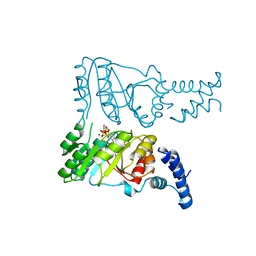 | | Crystal structure of FlhF complexed with GTP/Mg(2+) | | Descriptor: | Flagellar biosynthesis protein flhF, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5AFF
 
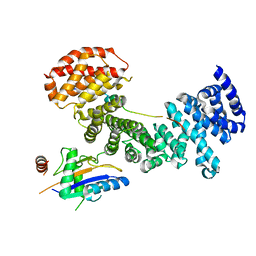 | | Symportin 1 chaperones 5S RNP assembly during ribosome biogenesis by occupying an essential rRNA binding site | | Descriptor: | RIBOSOMAL PROTEIN L11, RIBOSOMAL PROTEIN L5, SYMPORTIN 1 | | Authors: | Calvino, F.R, Kharde, S, Wild, K, Bange, G, Sinning, I. | | Deposit date: | 2015-01-21 | | Release date: | 2015-04-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.398 Å) | | Cite: | Symportin 1 Chaperones 5S Rnp Assembly During Ribosome Biogenesis by Occupying an Essential Rrna-Binding Site.
Nat.Commun., 6, 2015
|
|
2PX0
 
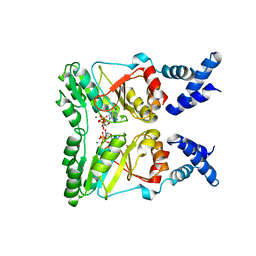 | | Crystal structure of FlhF complexed with GMPPNP/Mg(2+) | | Descriptor: | Flagellar biosynthesis protein flhF, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2Q8K
 
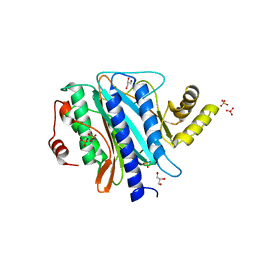 | | The crystal structure of Ebp1 | | Descriptor: | GLYCEROL, Proliferation-associated protein 2G4, SULFATE ION | | Authors: | Kowalinski, E, Bange, G, Wild, K, Sinning, I. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Ebp1 reveals a methionine aminopeptidase fold as binding platform for multiple interactions.
Febs Lett., 581, 2007
|
|
2QY9
 
 | |
4P3F
 
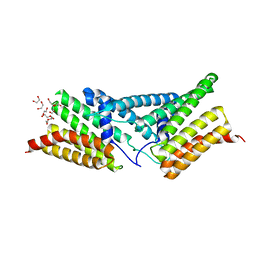 | | Structure of the human SRP68-RBD | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal recognition particle subunit SRP68 | | Authors: | Grotwinkel, J.T, Wild, K, Sinning, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
4P3E
 
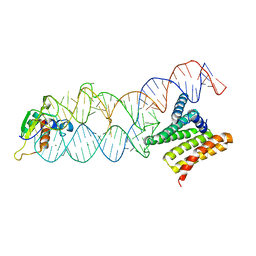 | | Structure of the human SRP S domain | | Descriptor: | MAGNESIUM ION, SRP RNA (124-mer), Signal recognition particle 19 kDa protein, ... | | Authors: | Grotwinkel, J.T, Wild, K, Sinning, I. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | SRP RNA remodeling by SRP68 explains its role in protein translocation.
Science, 344, 2014
|
|
5BY8
 
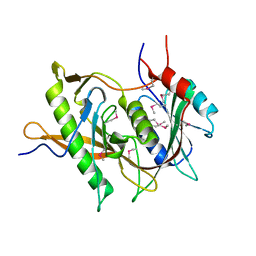 | | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis | | Descriptor: | Rpf2, Rrs1 | | Authors: | Kharde, S, Calvino, F.R, Gumiero, A, Wild, K, Sinning, I. | | Deposit date: | 2015-06-10 | | Release date: | 2015-07-08 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis.
Nucleic Acids Res., 43, 2015
|
|
4P3G
 
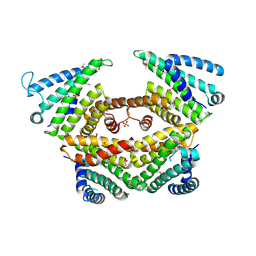 | |
5CK4
 
 | |
5CK5
 
 | |
5CK3
 
 | | Signal recognition particle receptor SRb-GTP/SRX complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jadhav, B.R, Wild, K, Sinning, I. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Switch Cycle of SR beta as Ancestral Eukaryotic GTPase Associated with Secretory Membranes.
Structure, 23, 2015
|
|
1QZW
 
 | | Crystal structure of the complete core of archaeal SRP and implications for inter-domain communication | | Descriptor: | 7S RNA, Signal recognition 54 kDa protein | | Authors: | Rosendal, K.R, Wild, K, Montoya, G, Sinning, I. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QZX
 
 | |
