3KTV
 
 | | Crystal structure of the human SRP19/S-domain SRP RNA complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KTW
 
 | | Crystal structure of the SRP19/S-domain SRP RNA complex of Sulfolobus solfataricus | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LPZ
 
 | | Crystal structure of C. therm. Get4 | | Descriptor: | Uncharacterized protein | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis.
Febs Lett., 584, 2010
|
|
3MAL
 
 | | Crystal structure of the SDF2-like protein from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stromal cell-derived factor 2-like protein | | Authors: | Ravaud, S, Radzimanowski, J, Sinning, I. | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Arabidopsis stromal-derived Factor2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum.
J.Biol.Chem., 285, 2010
|
|
3MIX
 
 | | Crystal structure of the cytosolic domain of B. subtilis FlhA | | Descriptor: | Flagellar biosynthesis protein flhA | | Authors: | Bange, G, Kuemmerer, N, Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2010-04-12 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4GMO
 
 | | Crystal structure of Syo1 | | Descriptor: | Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMQ
 
 | | Ribosome-binding domain of Zuo1 | | Descriptor: | Putative ribosome associated protein | | Authors: | Kopp, J, Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GMN
 
 | | Structural basis of Rpl5 recognition by Syo1 | | Descriptor: | 60S ribosomal protein l5-like protein, Putative uncharacterized protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
3SYN
 
 | | Crystal structure of FlhF in complex with its activator | | Descriptor: | ALUMINUM FLUORIDE, ATP-binding protein YlxH, Flagellar biosynthesis protein flhF, ... | | Authors: | Bange, G, Kuemmerer, N, Wild, K, Sinning, I. | | Deposit date: | 2011-07-18 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Structural basis for the molecular evolution of SRP-GTPase activation by protein.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4GNI
 
 | | Structure of the Ssz1 ATPase bound to ATP and Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative heat shock protein | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2012-08-17 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4IPA
 
 | | Structure of a thermophilic Arx1 | | Descriptor: | Putative curved DNA-binding protein, SULFATE ION | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2013-01-09 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Consistent mutational paths predict eukaryotic thermostability.
BMC Evol Biol, 13, 2013
|
|
3ZRI
 
 | |
3ZRJ
 
 | | Complex of ClpV N-domain with VipB peptide | | Descriptor: | 1,2-ETHANEDIOL, CLPB PROTEIN, VIPB | | Authors: | Lenherr, E.D, Kopp, J, Sinning, I. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Basis for the Unique Role of the Aaa+ Chaperone Clpv in Type Vi Protein Secretion.
J.Biol.Chem., 286, 2011
|
|
6QTB
 
 | | Crystal structure of Rea1-MIDAS/Ytm1-UBL complex from Chaetomium thermophilum | | Descriptor: | GLYCEROL, MAGNESIUM ION, Midasin,Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
6QT8
 
 | | Crystal structure of Rea1-MIDAS domain from Chaetomium thermophilum | | Descriptor: | GLYCEROL, IODIDE ION, Midasin, ... | | Authors: | Ahmed, Y.L, Thoms, M, Hurt, E, Sinning, I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of Rea1-MIDAS bound to its ribosome assembly factor ligands resembling integrin-ligand-type complexes.
Nat Commun, 10, 2019
|
|
6RYI
 
 | | WUS-HD bound to G-Box DNA | | Descriptor: | DNA (5'-D(P*CP*CP*CP*AP*TP*CP*AP*CP*GP*TP*GP*AP*CP*GP*AP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*TP*CP*AP*CP*GP*TP*GP*AP*TP*GP*GP*G)-3'), Protein WUSCHEL | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6RYD
 
 | | WUS-HD bound to TGAA DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*GP*TP*AP*TP*GP*AP*AP*TP*GP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*CP*AP*TP*TP*CP*AP*TP*AP*CP*AP*CP*T)-3'), MAGNESIUM ION, ... | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6RYL
 
 | | WUS-HD bound to TAAT DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*AP*AP*CP*CP*CP*AP*TP*TP*AP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*TP*AP*AP*TP*GP*GP*GP*TP*TP*GP*TP*G)-3'), Protein WUSCHEL | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6RY3
 
 | | Structure of the WUS homeodomain | | Descriptor: | ACETYL GROUP, Protein WUSCHEL, SULFATE ION | | Authors: | Sloan, J.J, Wild, K, Sinning, I. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Structural basis for the complex DNA binding behavior of the plant stem cell regulator WUSCHEL.
Nat Commun, 11, 2020
|
|
6SO5
 
 | | Homo sapiens WRB/CAML heterotetramer in complex with a TRC40 dimer | | Descriptor: | ATPase ASNA1, Calcium signal-modulating cyclophilin ligand, Tail-anchored protein insertion receptor WRB, ... | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Flemming, D, Sinning, I. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-09 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
Mol.Cell, 80, 2020
|
|
6SR6
 
 | | Crystal structure of the RAC core with a pseudo substrate bound to Ssz1 SBD | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Putative heat shock protein, ... | | Authors: | Valentin Gese, G, Lapouge, K, Kopp, J, Sinning, I. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The ribosome-associated complex RAC serves in a relay that directs nascent chains to Ssb.
Nat Commun, 11, 2020
|
|
6SXO
 
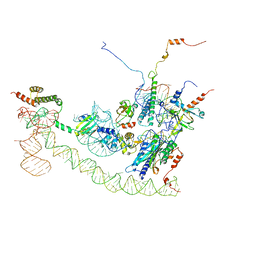 | | Cryo-EM structure of the human Ebp1-ribosome complex | | Descriptor: | 28S ribosomal RNA including ES27L-B (2839-3265), 5.8S ribosomal RNA, 60S ribosomal protein L19, ... | | Authors: | Wild, K, Aleksic, M, Pfeffer, M, Sinning, I. | | Deposit date: | 2019-09-26 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | MetAP-like Ebp1 occupies the human ribosomal tunnel exit and recruits flexible rRNA expansion segments.
Nat Commun, 11, 2020
|
|
3BS6
 
 | |
6YFV
 
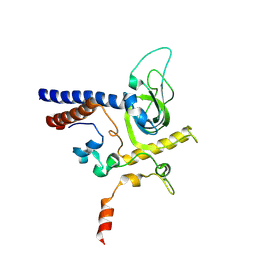 | | Crystal structure of Mtr4-Red1 minimal complex from Chaetomium thermophilum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP dependent RNA helicase (Dob1)-like protein, Red1, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-26 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
6YGU
 
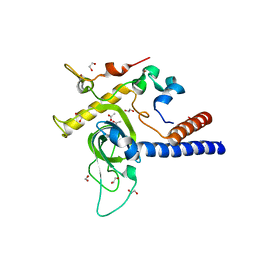 | | Crystal structure of the minimal Mtr4-Red1 complex (single chain) from Chaetomium thermophilum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ATP dependent RNA helicase (Dob1)-like protein, ... | | Authors: | Dobrev, N, Ahmed, Y.L, Sinning, I. | | Deposit date: | 2020-03-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The zinc-finger protein Red1 orchestrates MTREC submodules and binds the Mtl1 helicase arch domain.
Nat Commun, 12, 2021
|
|
