5D6C
 
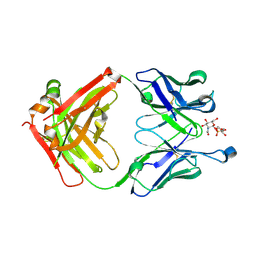 | | Structure of 4497 Fab bound to synthetic wall teichoic acid fragment | | Descriptor: | 4-O-[2-acetamido-2-deoxy-beta-D-glucopyranosyl]-5-O-phosphono-D-ribitol, 4497 antibody IgG1 (VH and CH1), 4497 antibody IgK (VL and CL), ... | | Authors: | Lupardus, P.J, Fong, R. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel antibody-antibiotic conjugate eliminates intracellular S. aureus.
Nature, 527, 2015
|
|
5KUT
 
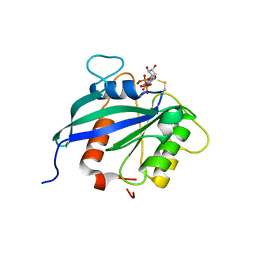 | | hMiro2 C-terminal GTPase domain, GDP-bound | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 2 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSY
 
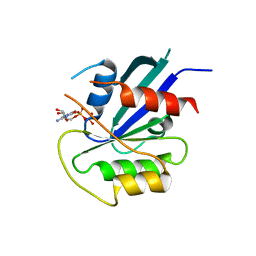 | | hMiro1 C-domain GDP Complex P41212 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSZ
 
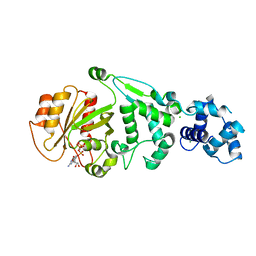 | | hMiro EF hand and cGTPase domains in the GMPPCP-bound state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mitochondrial Rho GTPase 1, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KTY
 
 | | hMiro EF hand and cGTPase domains, GDP and Ca2+ bound state | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5YUK
 
 | |
5KSP
 
 | | hMiro1 C-domain GDP Complex C2221 Crystal Form | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5YUI
 
 | | CO2 release in human carbonic anhydrase II crystals: reveal histidine 64 and solvent dynamics | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Park, S.Y, McKenna, R. | | Deposit date: | 2017-11-22 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Tracking solvent and protein movement during CO2 release in carbonic anhydrase II crystals.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5KU1
 
 | | hMiro1 EF hand and cGTPase domains in the GDP-bound state | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSO
 
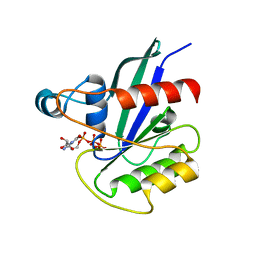 | | hMiro1 C-domain GDP-Pi Complex P3121 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1, PHOSPHATE ION | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5YUJ
 
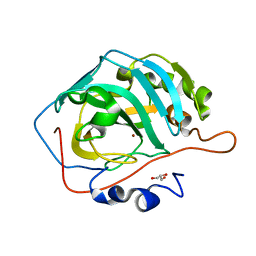 | |
7JL3
 
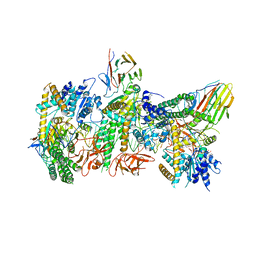 | | Cryo-EM structure of RIG-I:dsRNA filament in complex with RIPLET PrySpry domain (trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL0
 
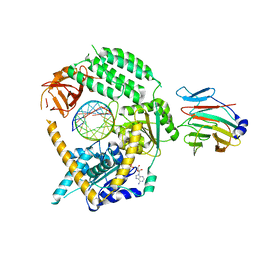 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL1
 
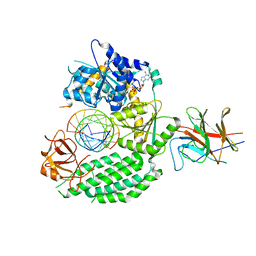 | | Cryo-EM structure of RIG-I:dsRNA in complex with RIPLET PrySpry domain (monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL4
 
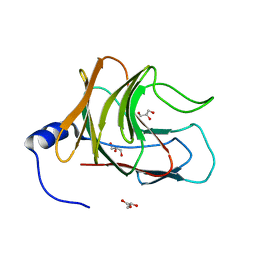 | | Crystal structure of TRIM65 PSpry domain | | Descriptor: | GLYCEROL, Tripartite motif-containing protein 65 | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
7JL2
 
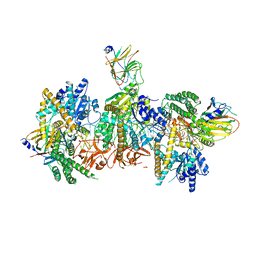 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
3P8D
 
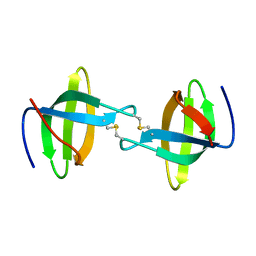 | | Crystal structure of the second Tudor domain of human PHF20 (homodimer form) | | Descriptor: | Medulloblastoma antigen MU-MB-50.72 | | Authors: | Cui, G, Lee, J, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2010-10-13 | | Release date: | 2011-06-22 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5Z2E
 
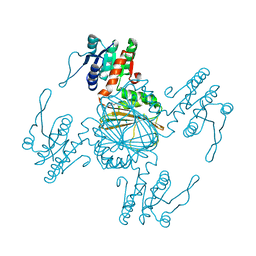 | |
5Z2F
 
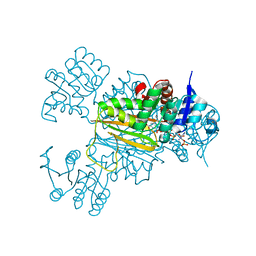 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
5Z2D
 
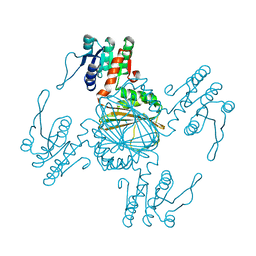 | |
6JCG
 
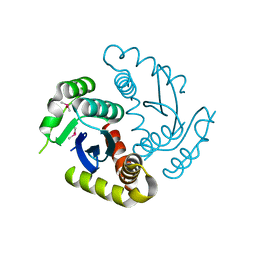 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6JCF
 
 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | Descriptor: | CACODYLATE ION, Integrase | | Authors: | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
7MY2
 
 | |
7MY3
 
 | |
6IG7
 
 | |
