4OGL
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymine at 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymine at 1.25 A resolution
Crystallogr. Rep., 2016
|
|
4OEH
 
 | | X-ray Structure of Uridine Phosphorylase from Vibrio cholerae Complexed with Uracil at 1.91 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases.
To Be Published, Crystallogr. Rep., 2016
|
|
5LH0
 
 | | Low dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LN0
 
 | | Low dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH7
 
 | | High dose Thaumatin - 760-800 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LHV
 
 | | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with uridine and sulfate ion at 1.29 A resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Balaev, V.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2016-07-13 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with uridine and sulfate ion at 1.29 A resolution
To Be Published
|
|
5LH3
 
 | | High dose Thaumatin - 0-40 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LOK
 
 | | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.11 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Dontsova, M.V, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.109 Å) | | Cite: | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.11 A resolution
To Be Published
|
|
5LH6
 
 | | High dose Thaumatin - 360-400 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH1
 
 | | Low dose Thaumatin - 360-400 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LH5
 
 | | High dose Thaumatin - 40-80 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5LMH
 
 | | High dose Thaumatin - 160-200 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
5M2T
 
 | | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with uridine at 1.03 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | X-ray structure of uridine phosphorylase from Vibrio cholerae in complex with uridine at 1.03 A resolution
To Be Published
|
|
6ELY
 
 | | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.84 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-N-Furfurylcytosine, ... | | Authors: | Ahmad, M.S, Rasheed, S, Falke, S, Khaliq, B, Perbandt, M, Choudhary, M.I, Markiewicz, W.T, Barciszewski, J, Betzel, C. | | Deposit date: | 2017-09-30 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.85 angstrom Resolution.
Med Chem, 14, 2018
|
|
4YEK
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with thymidine | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6EYP
 
 | | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Prokofev, I.I, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Lashkov, A.A. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | X-ray structure of the unliganded uridine phosphorylase from Vibrio cholerae at 1.22A
To Be Published
|
|
4YYY
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with uridine | | Descriptor: | CITRIC ACID, TRIETHYLENE GLYCOL, Thymidine phosphorylase, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-03-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4ZXG
 
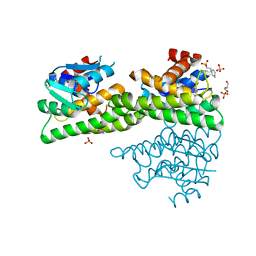 | | Ligandin binding site of PfGST | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Perbandt, M, Eberle, R, Betzel, C. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structures of Plasmodium falciparum GST complexes provide novel insights into the dimer-tetramer transition and a novel ligand-binding site.
J.Struct.Biol., 191, 2015
|
|
6HMZ
 
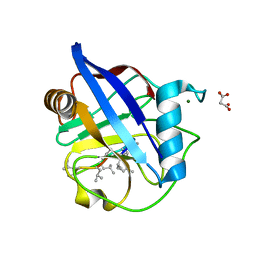 | | Crystal Structure of a Single-Domain Cyclophilin from Brassica napus Phloem Sap | | Descriptor: | Cyclosporin, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Falke, S, Hanhart, P, Garbe, M, Thiess, M, Betzel, C, Kehr, J. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Enzyme activity and structural features of three single-domain phloem cyclophilins from Brassica napus.
Sci Rep, 9, 2019
|
|
5C80
 
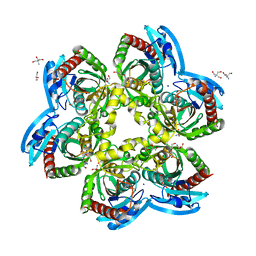 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-06-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases
Crystallography Reports, 61, 2016
|
|
5D73
 
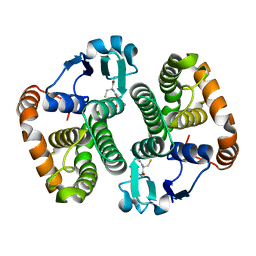 | | Structure of Wuchereria bancrofti pi-class glutathione S-transferase | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Prince, P.R, Sakthidevi, M, Madhumathi, J, Perbandt, M, Betzel, C, Kaliraj, P. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | STRUCTURE OF WUCHERERIA BANCROFTI PI-CLASS GLUTATHIONE S-TRANSFERASE
TO BE PUBLISHED
|
|
1LNL
 
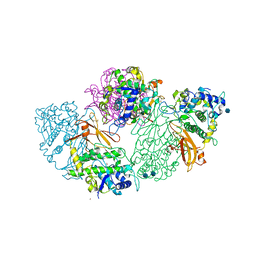 | | Structure of deoxygenated hemocyanin from Rapana thomasiana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, hemocyanin | | Authors: | Perbandt, M, Guthoehrlein, E.W, Rypniewski, W, Idakieva, K, Stoeva, S, Voelter, W, Genov, N, Betzel, C. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Biochemistry, 42, 2003
|
|
2V6W
 
 | | tRNASer acceptor stem: Conformation and hydration of a microhelix in a crystal structure at 1.8 Angstrom resolution | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*AP)-3', 5'-R(*UP*CP*UP*CP*UP*CP*CP)-3' | | Authors: | Foerster, C, Brauer, A.B.E, Brode, S, Fuerste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trnaser Acceptor Stem: Conformation and Hydration of a Microhelix in a Crystal Structure at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2W89
 
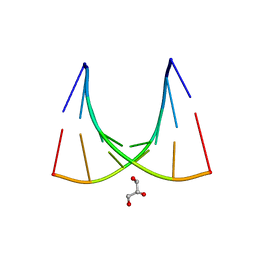 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
2HNL
 
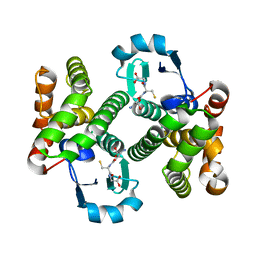 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1 | | Authors: | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
