2N7X
 
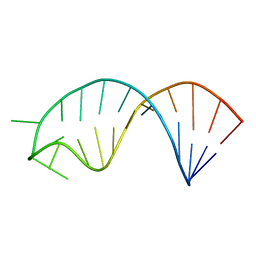 | | Solution structure of microRNA 20b pre-element | | Descriptor: | RNA (5'-R(*GP*GP*UP*AP*GP*UP*UP*UP*UP*GP*GP*CP*AP*UP*GP*AP*CP*UP*CP*UP*AP*CP*C)-3') | | Authors: | Yang, F, Chen, Y, Varani, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer.
Nucleic Acids Res., 44, 2016
|
|
2N6S
 
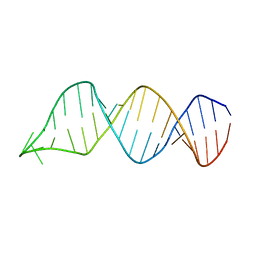 | |
2OSQ
 
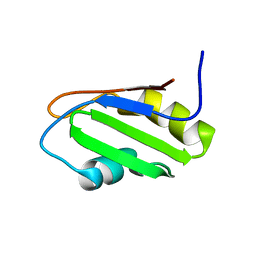 | | NMR Structure of RRM-1 of Yeast NPL3 Protein | | Descriptor: | Nucleolar protein 3 | | Authors: | Deka, P, Bucheli, M, Skrisovska, L, Allain, F.H, Moore, C, Buratowski, S, Varani, G. | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast SR protein Npl3 and Interaction with mRNA 3'-end processing signals.
J.Mol.Biol., 375, 2008
|
|
2OSR
 
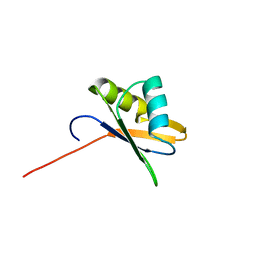 | | NMR Structure of RRM-2 of Yeast NPL3 Protein | | Descriptor: | Nucleolar protein 3 | | Authors: | Deka, P, Bucheli, M, Skrisovska, L, Allain, F.H, Moore, C, Buratowski, S, Varani, G. | | Deposit date: | 2007-02-06 | | Release date: | 2007-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the yeast SR protein Npl3 and Interaction with mRNA 3'-end processing signals.
J.Mol.Biol., 375, 2008
|
|
2MZY
 
 | | 1H, 13C, and 15N Chemical Shift Assignments and structure of Probable Fe(2+)-trafficking protein from Burkholderia pseudomallei 1710b. | | Descriptor: | Probable Fe(2+)-trafficking protein | | Authors: | Tang, C, Yang, F, Barnwal, R, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Probable Fe(2+)-trafficking protein
To be Published
|
|
2N3S
 
 | |
2N57
 
 | |
2N6W
 
 | |
2MRC
 
 | |
2Y78
 
 | | Crystal structure of BPSS1823, a Mip-like chaperone from Burkholderia pseudomallei | | Descriptor: | CHLORIDE ION, GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | Authors: | Norville, I.H, O'Shea, K, Sarkar-Tyson, M, Harmer, N.J. | | Deposit date: | 2011-01-28 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | The Structure of a Burkholderia Pseudomallei Immunophilin-Inhibitor Complex Reveals New Approaches to Antimicrobial Development
Biochem.J., 437, 2011
|
|
2GJH
 
 | |
2GJF
 
 | |
2NS4
 
 | |
1AJU
 
 | |
1AKX
 
 | |
