7FES
 
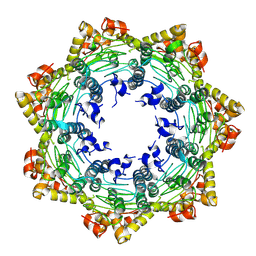 | | Cryo-EM structure of apo BsClpP at pH 4.2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FEQ
 
 | | Cryo-EM structure of apo BsClpP at pH 6.5 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
3NIH
 
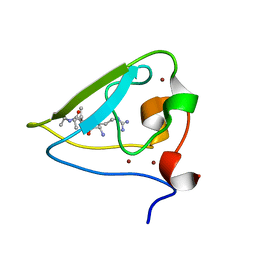 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIL
 
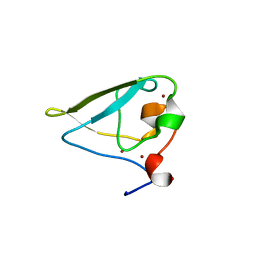 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
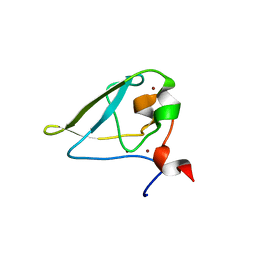 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NII
 
 | | The structure of UBR box (KIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide KIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIM
 
 | | The structure of UBR box (RRAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RRAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIJ
 
 | | The structure of UBR box (HIAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide HIAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIN
 
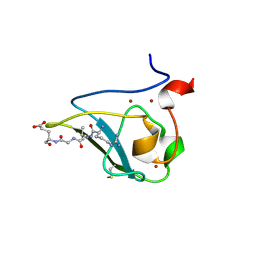 | | The structure of UBR box (RLGES) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RLGES, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIT
 
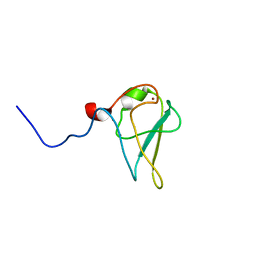 | | The structure of UBR box (native1) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3PO0
 
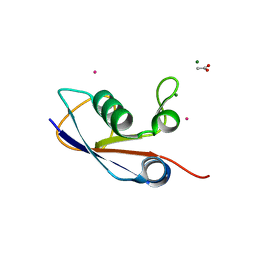 | | Crystal structure of SAMP1 from Haloferax volcanii | | Descriptor: | ACETATE ION, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Jeong, Y.J, Jeong, B.-C, Song, H.K. | | Deposit date: | 2010-11-21 | | Release date: | 2011-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
3RUJ
 
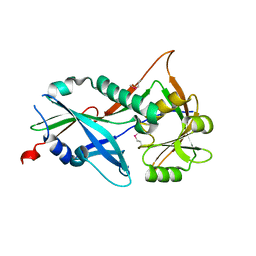 | |
3RUI
 
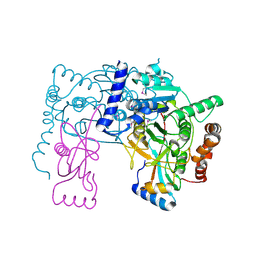 | | Crystal structure of Atg7C-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Hong, S.B, Kim, B.W, Song, H.K. | | Deposit date: | 2011-05-05 | | Release date: | 2011-11-23 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SL7
 
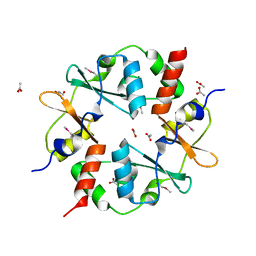 | | Crystal structure of CBS-pair protein, CBSX2 from Arabidopsis thaliana | | Descriptor: | ACETATE ION, CBS domain-containing protein CBSX2, GLYCEROL | | Authors: | Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2011-06-24 | | Release date: | 2011-11-09 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Single cystathionine beta-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis
Plant Cell, 23, 2011
|
|
5HZY
 
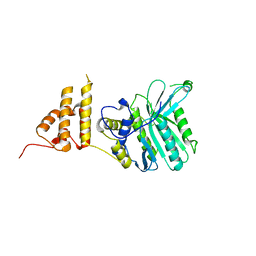 | | Crystal structure of the legionella pneumophila effector protein RavZ - P6322 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-02-03 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
5IO3
 
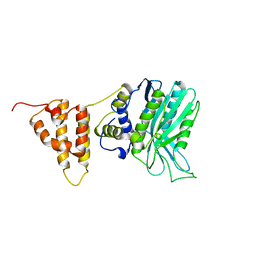 | | Crystal structure of the legionella pneumophila effector protein RavZ - I422 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
7XV2
 
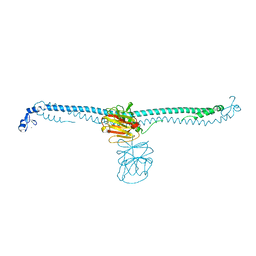 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-05-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XZ1
 
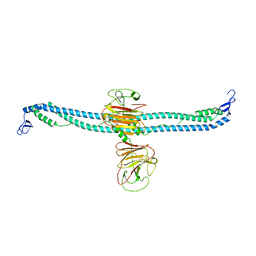 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XZ0
 
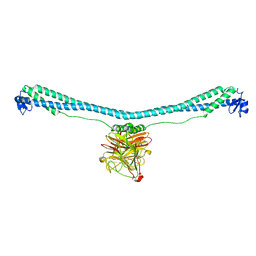 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XYZ
 
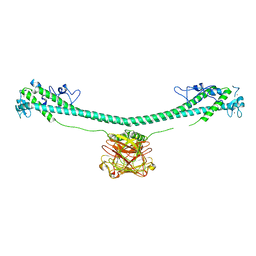 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.62 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XZ2
 
 | | TRIM E3 ubiquitin ligase | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XYY
 
 | | TRIM E3 ubiquitin ligase WT | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Park, S.H, Song, H.K. | | Deposit date: | 2022-06-02 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (7.1 Å) | | Cite: | Structure and activation of the RING E3 ubiquitin ligase TRIM72 on the membrane.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6KHZ
 
 | | p62/SQSTM1 ZZ domain with Gly-peptide | | Descriptor: | Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2019-07-16 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Use of the LC3B-fusion technique for biochemical and structural studies of proteins involved in the N-degron pathway.
J.Biol.Chem., 295, 2020
|
|
6KGI
 
 | | RLGS-yUbr1 Ubr box | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Heo, J, Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2019-07-11 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Use of the LC3B-fusion technique for biochemical and structural studies of proteins involved in the N-degron pathway.
J.Biol.Chem., 295, 2020
|
|
