5D2Q
 
 | |
5D23
 
 | |
6WJ6
 
 | | Cryo-EM structure of apo-Photosystem II from Synechocystis sp. PCC 6803 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gisriel, C.J. | | Deposit date: | 2020-04-12 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM Structure of Monomeric Photosystem II from Synechocystis sp. PCC 6803 Lacking the Water-Oxidation Complex
Joule, 2020
|
|
3SA0
 
 | | Complex of ERK2 with norathyriol | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION, norathyriol | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5947 Å) | | Cite: | Norathyriol Suppresses Skin Cancers Induced by Solar Ultraviolet Radiation by Targeting ERK Kinases.
Cancer Res., 72, 2012
|
|
8H3Z
 
 | |
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
5KG8
 
 | |
6EQT
 
 | |
6UFR
 
 | | Structure of recombinantly assembled E46K alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Eisenberg, D.S, Boyer, D.R, Sawaya, M.R, Li, B, Jiang, L. | | Deposit date: | 2019-09-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2FFL
 
 | |
3GOD
 
 | |
376D
 
 | | A ZIPPER-LIKE DNA DUPLEX D(GCGAAAGCT) | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*(CBR)P*GP*AP*AP*AP*GP*CP*T)-3') | | Authors: | Cruse, W.B.T, Shepard, W, Prange, T, delalFortelle, E, Fourme, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 A resolution.
Structure, 6, 1998
|
|
1GRZ
 
 | |
4N0S
 
 | | Complex of ERK2 with caffeic acid | | Descriptor: | CAFFEIC ACID, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Kurinov, I, Malakhova, M. | | Deposit date: | 2013-10-02 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7992 Å) | | Cite: | Caffeic Acid Directly Targets ERK1/2 to Attenuate Solar UV-Induced Skin Carcinogenesis.
Cancer Prev Res (Phila), 7, 2014
|
|
6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | Descriptor: | HIV-1 CA | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6EC2
 
 | | Structure of HIV-1 CA 1/3-hexamer | | Descriptor: | ACETATE ION, Capsid protein p24 | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
6ECO
 
 | | Hexamer-2-Foldon HIV-1 capsid platform | | Descriptor: | HIV-1 capsid platform protein | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
4ZT9
 
 | |
4ZT0
 
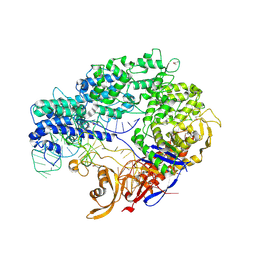 | |
7E9G
 
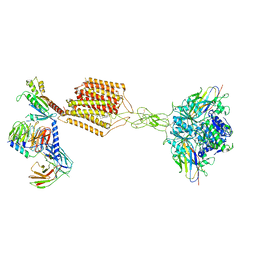 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7E9H
 
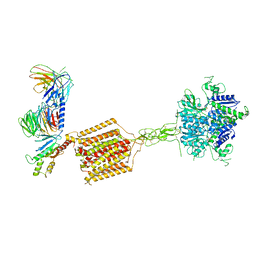 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
4OGC
 
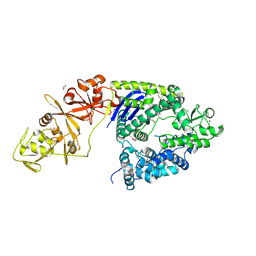 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
4OGE
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | HNH endonuclease domain protein, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
