5NJQ
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 4ET8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5NJP
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
5MND
 
 | |
5MJL
 
 | | Single-shot pink beam serial crystallography: Proteinase K | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21013784 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5MJP
 
 | | Multi-bunch pink beam serial crystallography: Phycocyanin (One chip) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5MJM
 
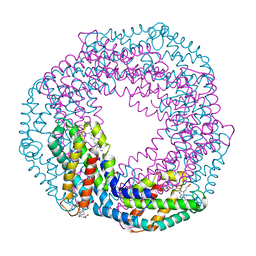 | | Single-shot pink beam serial crystallography: Phycocyanin (Five chips merged) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5MJQ
 
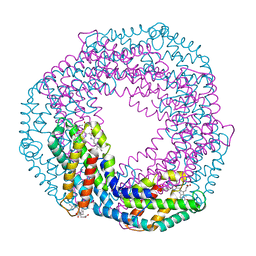 | | Single-shot pink beam serial crystallography: Phycocyanin (One chip) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5O7M
 
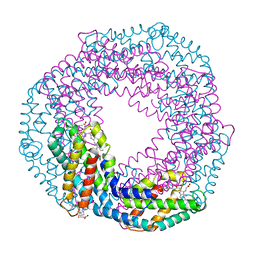 | | Single-shot pink beam serial crystallography: Phycocyanin (One chip, chip_1) | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V, Sarrou, I. | | Deposit date: | 2017-06-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
