4YCL
 
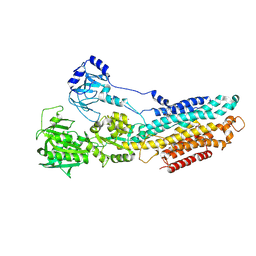 | | Crystal structure of the SR CA2+-ATPASE with bound CPA | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ogawa, H, Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2H5L
 
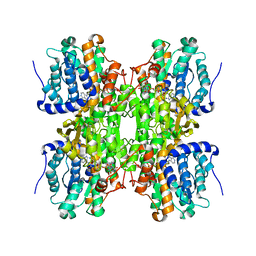 | | S-Adenosylhomocysteine hydrolase containing NAD and 3-deaza-D-eritadenine | | Descriptor: | (2R,3R)-4-(4-AMINO-1H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-2,3-DIHYDROXYBUTANOIC ACID, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, T, Komoto, J, Takusagawa, F. | | Deposit date: | 2006-05-26 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and function of eritadenine and its 3-deaza analogues: Potent inhibitors of S-adenosylhomocysteine hydrolase and hypocholesterolemic agents.
Biochem.Pharm., 73, 2007
|
|
1D2G
 
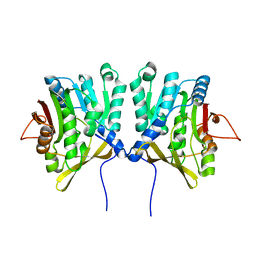 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE FROM RAT LIVER | | Descriptor: | GLYCINE N-METHYLTRANSFERASE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-08 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1D2C
 
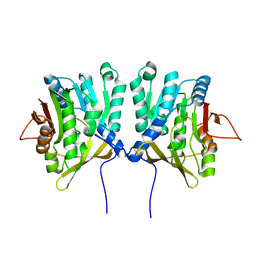 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1D2H
 
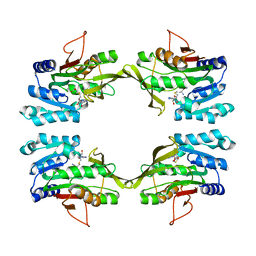 | | CRYSTAL STRUCTURE OF R175K MUTANT GLYCINE N-METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLHOMOCYSTEINE | | Descriptor: | GLYCINE N-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Huang, Y, Komoto, J, Takusagawa, F, Konishi, K, Takata, Y. | | Deposit date: | 1999-10-11 | | Release date: | 1999-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
1KY4
 
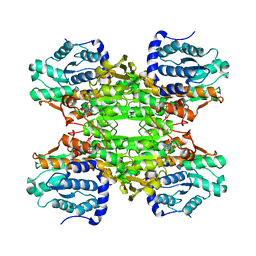 | |
1KY5
 
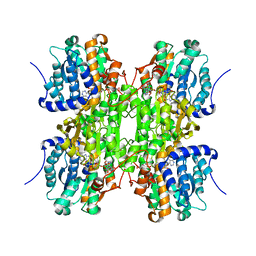 | | D244E mutant S-Adenosylhomocysteine hydrolase refined with noncrystallographic restraints | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3'-OXO-ADENOSINE, S-adenosylhomocysteine hydrolase | | Authors: | Takata, Y, Takusagawa, F. | | Deposit date: | 2002-02-03 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic Mechanism of S-adenosylhomocysteine hydrolase. Site-directed
mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190.
J.Biol.Chem., 277, 2002
|
|
1P1C
 
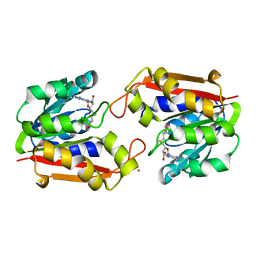 | | Guanidinoacetate Methyltransferase with Gd ion | | Descriptor: | GADOLINIUM ION, Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Takusagawa, F. | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Monoclinic guanidinoacetate methyltransferase and gadolinium ion-binding characteristics.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P1B
 
 | | Guanidinoacetate methyltransferase | | Descriptor: | Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Takusagawa, F. | | Deposit date: | 2003-04-12 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monoclinic guanidinoacetate methyltransferase and gadolinium ion-binding characteristics.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1XCJ
 
 | | Guanidinoacetate methyltransferase containing S-adenosylhomocysteine and guanidinoacetate | | Descriptor: | GUANIDINO ACETATE, Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Yamada, T, Takata, Y, Takusagawa, F. | | Deposit date: | 2004-09-02 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of guanidinoacetate methyltransferase: crystal structures of guanidinoacetate methyltransferase ternary complexes.
Biochemistry, 43, 2004
|
|
1XCL
 
 | | Guanidinoacetate methyltransferase containing S-adenosylhomocysteine and guanidine | | Descriptor: | GUANIDINE, Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Yamada, T, Takata, Y, Takusagawa, F. | | Deposit date: | 2004-09-02 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of guanidinoacetate methyltransferase: crystal structures of guanidinoacetate methyltransferase ternary complexes.
Biochemistry, 43, 2004
|
|
1IWO
 
 | |
2AGV
 
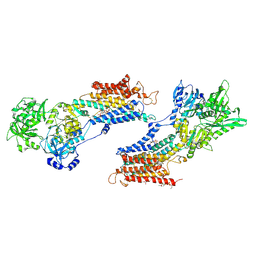 | | Crystal structure of the SR CA2+-ATPASE with BHQ and TG | | Descriptor: | 2,5-DITERT-BUTYLBENZENE-1,4-DIOL, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Obara, K, Norimatsu, Y. | | Deposit date: | 2005-07-27 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inaugural Article: Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1WPG
 
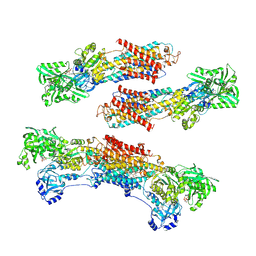 | | Crystal structure of the SR CA2+-ATPase with MGF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T. | | Deposit date: | 2004-09-02 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
2DQS
 
 | |
2EAT
 
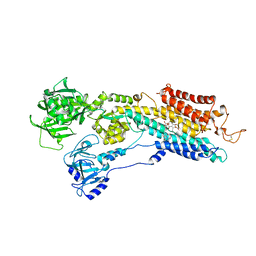 | | Crystal structure of the SR CA2+-ATPASE with bound CPA and TG | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EAU
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA in the presence of curcumin | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, PHOSPHATIDYLETHANOLAMINE, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2EAR
 
 | | P21 crystal of the SR CA2+-ATPase with bound TG | | Descriptor: | OCTANOIC ACID [3S-[3ALPHA, 3ABETA, 4ALPHA, ... | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1KHH
 
 | |
1NBI
 
 | |
1NBH
 
 | |
1VFP
 
 | |
3ONE
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3ONF
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with cordycepin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3'-DEOXYADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OND
 
 | | Crystal structure of Lupinus luteus S-adenosyl-L-homocysteine hydrolase in complex with adenosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Brzezinski, K, Jaskolski, M. | | Deposit date: | 2010-08-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | High-resolution structures of complexes of plant S-adenosyl-L-homocysteine hydrolase (Lupinus luteus).
Acta Crystallogr.,Sect.D, 68, 2012
|
|
